mirror of
https://github.com/lancedb/lancedb.git
synced 2025-12-23 13:29:57 +00:00
Compare commits
45 Commits
python-v0.
...
python-v0.
| Author | SHA1 | Date | |
|---|---|---|---|
|
|
ff5bbfdd4c | ||
|
|
694ca30c7c | ||
|
|
b2317c904d | ||
|
|
613f3063b9 | ||
|
|
5d2cd7fb2e | ||
|
|
a88e9bb134 | ||
|
|
9c1adff426 | ||
|
|
f9d5fa88a1 | ||
|
|
4db554eea5 | ||
|
|
101066788d | ||
|
|
c4135d9d30 | ||
|
|
ec39d98571 | ||
|
|
0cb37f0e5e | ||
|
|
24e3507ee2 | ||
|
|
2bdf0a02f9 | ||
|
|
32123713fd | ||
|
|
d5a01ffe7b | ||
|
|
e01045692c | ||
|
|
a62f661d90 | ||
|
|
4769d8eb76 | ||
|
|
d07d7a5980 | ||
|
|
8d2ff7b210 | ||
|
|
61c05b51a0 | ||
|
|
7801ab9b8b | ||
|
|
d297da5a7e | ||
|
|
6af69b57ad | ||
|
|
a062a92f6b | ||
|
|
277b753fd8 | ||
|
|
f78b7863f6 | ||
|
|
e7d824af2b | ||
|
|
02f1ec775f | ||
|
|
7b6d3f943b | ||
|
|
676876f4d5 | ||
|
|
fbfe2444a8 | ||
|
|
9555efacf9 | ||
|
|
513926960d | ||
|
|
cc507ca766 | ||
|
|
492d0328fe | ||
|
|
374c1e7aba | ||
|
|
30047a5566 | ||
|
|
85ccf9e22b | ||
|
|
0255221086 | ||
|
|
4ee229490c | ||
|
|
93e24f23af | ||
|
|
8f141e1e33 |
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.7.1"
|
current_version = "0.9.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
|
|||||||
48
.github/workflows/java.yml
vendored
48
.github/workflows/java.yml
vendored
@@ -3,6 +3,8 @@ on:
|
|||||||
push:
|
push:
|
||||||
branches:
|
branches:
|
||||||
- main
|
- main
|
||||||
|
paths:
|
||||||
|
- java/**
|
||||||
pull_request:
|
pull_request:
|
||||||
paths:
|
paths:
|
||||||
- java/**
|
- java/**
|
||||||
@@ -21,9 +23,42 @@ env:
|
|||||||
CARGO_INCREMENTAL: "0"
|
CARGO_INCREMENTAL: "0"
|
||||||
CARGO_BUILD_JOBS: "1"
|
CARGO_BUILD_JOBS: "1"
|
||||||
jobs:
|
jobs:
|
||||||
linux-build:
|
linux-build-java-11:
|
||||||
runs-on: ubuntu-22.04
|
runs-on: ubuntu-22.04
|
||||||
name: ubuntu-22.04 + Java 11 & 17
|
name: ubuntu-22.04 + Java 11
|
||||||
|
defaults:
|
||||||
|
run:
|
||||||
|
working-directory: ./java
|
||||||
|
steps:
|
||||||
|
- name: Checkout repository
|
||||||
|
uses: actions/checkout@v4
|
||||||
|
- uses: Swatinem/rust-cache@v2
|
||||||
|
with:
|
||||||
|
workspaces: java/core/lancedb-jni
|
||||||
|
- name: Run cargo fmt
|
||||||
|
run: cargo fmt --check
|
||||||
|
working-directory: ./java/core/lancedb-jni
|
||||||
|
- name: Install dependencies
|
||||||
|
run: |
|
||||||
|
sudo apt update
|
||||||
|
sudo apt install -y protobuf-compiler libssl-dev
|
||||||
|
- name: Install Java 11
|
||||||
|
uses: actions/setup-java@v4
|
||||||
|
with:
|
||||||
|
distribution: temurin
|
||||||
|
java-version: 11
|
||||||
|
cache: "maven"
|
||||||
|
- name: Java Style Check
|
||||||
|
run: mvn checkstyle:check
|
||||||
|
# Disable because of issues in lancedb rust core code
|
||||||
|
# - name: Rust Clippy
|
||||||
|
# working-directory: java/core/lancedb-jni
|
||||||
|
# run: cargo clippy --all-targets -- -D warnings
|
||||||
|
- name: Running tests with Java 11
|
||||||
|
run: mvn clean test
|
||||||
|
linux-build-java-17:
|
||||||
|
runs-on: ubuntu-22.04
|
||||||
|
name: ubuntu-22.04 + Java 17
|
||||||
defaults:
|
defaults:
|
||||||
run:
|
run:
|
||||||
working-directory: ./java
|
working-directory: ./java
|
||||||
@@ -47,20 +82,12 @@ jobs:
|
|||||||
java-version: 17
|
java-version: 17
|
||||||
cache: "maven"
|
cache: "maven"
|
||||||
- run: echo "JAVA_17=$JAVA_HOME" >> $GITHUB_ENV
|
- run: echo "JAVA_17=$JAVA_HOME" >> $GITHUB_ENV
|
||||||
- name: Install Java 11
|
|
||||||
uses: actions/setup-java@v4
|
|
||||||
with:
|
|
||||||
distribution: temurin
|
|
||||||
java-version: 11
|
|
||||||
cache: "maven"
|
|
||||||
- name: Java Style Check
|
- name: Java Style Check
|
||||||
run: mvn checkstyle:check
|
run: mvn checkstyle:check
|
||||||
# Disable because of issues in lancedb rust core code
|
# Disable because of issues in lancedb rust core code
|
||||||
# - name: Rust Clippy
|
# - name: Rust Clippy
|
||||||
# working-directory: java/core/lancedb-jni
|
# working-directory: java/core/lancedb-jni
|
||||||
# run: cargo clippy --all-targets -- -D warnings
|
# run: cargo clippy --all-targets -- -D warnings
|
||||||
- name: Running tests with Java 11
|
|
||||||
run: mvn clean test
|
|
||||||
- name: Running tests with Java 17
|
- name: Running tests with Java 17
|
||||||
run: |
|
run: |
|
||||||
export JAVA_TOOL_OPTIONS="$JAVA_TOOL_OPTIONS \
|
export JAVA_TOOL_OPTIONS="$JAVA_TOOL_OPTIONS \
|
||||||
@@ -83,3 +110,4 @@ jobs:
|
|||||||
-Djdk.reflect.useDirectMethodHandle=false \
|
-Djdk.reflect.useDirectMethodHandle=false \
|
||||||
-Dio.netty.tryReflectionSetAccessible=true"
|
-Dio.netty.tryReflectionSetAccessible=true"
|
||||||
JAVA_HOME=$JAVA_17 mvn clean test
|
JAVA_HOME=$JAVA_17 mvn clean test
|
||||||
|
|
||||||
|
|||||||
31
Cargo.toml
31
Cargo.toml
@@ -20,29 +20,30 @@ keywords = ["lancedb", "lance", "database", "vector", "search"]
|
|||||||
categories = ["database-implementations"]
|
categories = ["database-implementations"]
|
||||||
|
|
||||||
[workspace.dependencies]
|
[workspace.dependencies]
|
||||||
lance = { "version" = "=0.14.1", "features" = ["dynamodb"] }
|
lance = { "version" = "=0.16.1", "features" = ["dynamodb"] }
|
||||||
lance-index = { "version" = "=0.14.1" }
|
lance-index = { "version" = "=0.16.1" }
|
||||||
lance-linalg = { "version" = "=0.14.1" }
|
lance-linalg = { "version" = "=0.16.1" }
|
||||||
lance-testing = { "version" = "=0.14.1" }
|
lance-testing = { "version" = "=0.16.1" }
|
||||||
lance-datafusion = { "version" = "=0.14.1" }
|
lance-datafusion = { "version" = "=0.16.1" }
|
||||||
|
lance-encoding = { "version" = "=0.16.1" }
|
||||||
# Note that this one does not include pyarrow
|
# Note that this one does not include pyarrow
|
||||||
arrow = { version = "51.0", optional = false }
|
arrow = { version = "52.2", optional = false }
|
||||||
arrow-array = "51.0"
|
arrow-array = "52.2"
|
||||||
arrow-data = "51.0"
|
arrow-data = "52.2"
|
||||||
arrow-ipc = "51.0"
|
arrow-ipc = "52.2"
|
||||||
arrow-ord = "51.0"
|
arrow-ord = "52.2"
|
||||||
arrow-schema = "51.0"
|
arrow-schema = "52.2"
|
||||||
arrow-arith = "51.0"
|
arrow-arith = "52.2"
|
||||||
arrow-cast = "51.0"
|
arrow-cast = "52.2"
|
||||||
async-trait = "0"
|
async-trait = "0"
|
||||||
chrono = "0.4.35"
|
chrono = "0.4.35"
|
||||||
datafusion-physical-plan = "37.1"
|
datafusion-physical-plan = "40.0"
|
||||||
half = { "version" = "=2.4.1", default-features = false, features = [
|
half = { "version" = "=2.4.1", default-features = false, features = [
|
||||||
"num-traits",
|
"num-traits",
|
||||||
] }
|
] }
|
||||||
futures = "0"
|
futures = "0"
|
||||||
log = "0.4"
|
log = "0.4"
|
||||||
object_store = "0.9.0"
|
object_store = "0.10.2"
|
||||||
pin-project = "1.0.7"
|
pin-project = "1.0.7"
|
||||||
snafu = "0.7.4"
|
snafu = "0.7.4"
|
||||||
url = "2"
|
url = "2"
|
||||||
|
|||||||
22
README.md
22
README.md
@@ -44,26 +44,24 @@ LanceDB's core is written in Rust 🦀 and is built using <a href="https://githu
|
|||||||
|
|
||||||
**Javascript**
|
**Javascript**
|
||||||
```shell
|
```shell
|
||||||
npm install vectordb
|
npm install @lancedb/lancedb
|
||||||
```
|
```
|
||||||
|
|
||||||
```javascript
|
```javascript
|
||||||
const lancedb = require('vectordb');
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
const db = await lancedb.connect('data/sample-lancedb');
|
|
||||||
|
|
||||||
const table = await db.createTable({
|
const db = await lancedb.connect("data/sample-lancedb");
|
||||||
name: 'vectors',
|
const table = await db.createTable("vectors", [
|
||||||
data: [
|
|
||||||
{ id: 1, vector: [0.1, 0.2], item: "foo", price: 10 },
|
{ id: 1, vector: [0.1, 0.2], item: "foo", price: 10 },
|
||||||
{ id: 2, vector: [1.1, 1.2], item: "bar", price: 50 }
|
{ id: 2, vector: [1.1, 1.2], item: "bar", price: 50 },
|
||||||
]
|
], {mode: 'overwrite'});
|
||||||
})
|
|
||||||
|
|
||||||
const query = table.search([0.1, 0.3]).limit(2);
|
|
||||||
const results = await query.execute();
|
const query = table.vectorSearch([0.1, 0.3]).limit(2);
|
||||||
|
const results = await query.toArray();
|
||||||
|
|
||||||
// You can also search for rows by specific criteria without involving a vector search.
|
// You can also search for rows by specific criteria without involving a vector search.
|
||||||
const rowsByCriteria = await table.search(undefined).where("price >= 10").execute();
|
const rowsByCriteria = await table.query().where("price >= 10").toArray();
|
||||||
```
|
```
|
||||||
|
|
||||||
**Python**
|
**Python**
|
||||||
|
|||||||
@@ -18,4 +18,4 @@ docker run \
|
|||||||
-v $(pwd):/io -w /io \
|
-v $(pwd):/io -w /io \
|
||||||
--memory-swap=-1 \
|
--memory-swap=-1 \
|
||||||
lancedb-node-manylinux \
|
lancedb-node-manylinux \
|
||||||
bash ci/manylinux_node/build.sh $ARCH
|
bash ci/manylinux_node/build_vectordb.sh $ARCH
|
||||||
|
|||||||
@@ -4,9 +4,9 @@ ARCH=${1:-x86_64}
|
|||||||
|
|
||||||
# We pass down the current user so that when we later mount the local files
|
# We pass down the current user so that when we later mount the local files
|
||||||
# into the container, the files are accessible by the current user.
|
# into the container, the files are accessible by the current user.
|
||||||
pushd ci/manylinux_nodejs
|
pushd ci/manylinux_node
|
||||||
docker build \
|
docker build \
|
||||||
-t lancedb-nodejs-manylinux \
|
-t lancedb-node-manylinux-$ARCH \
|
||||||
--build-arg="ARCH=$ARCH" \
|
--build-arg="ARCH=$ARCH" \
|
||||||
--build-arg="DOCKER_USER=$(id -u)" \
|
--build-arg="DOCKER_USER=$(id -u)" \
|
||||||
--progress=plain \
|
--progress=plain \
|
||||||
@@ -17,5 +17,5 @@ popd
|
|||||||
docker run \
|
docker run \
|
||||||
-v $(pwd):/io -w /io \
|
-v $(pwd):/io -w /io \
|
||||||
--memory-swap=-1 \
|
--memory-swap=-1 \
|
||||||
lancedb-nodejs-manylinux \
|
lancedb-node-manylinux-$ARCH \
|
||||||
bash ci/manylinux_nodejs/build.sh $ARCH
|
bash ci/manylinux_node/build_lancedb.sh $ARCH
|
||||||
|
|||||||
@@ -4,7 +4,7 @@
|
|||||||
# range of linux distributions.
|
# range of linux distributions.
|
||||||
ARG ARCH=x86_64
|
ARG ARCH=x86_64
|
||||||
|
|
||||||
FROM quay.io/pypa/manylinux2014_${ARCH}
|
FROM quay.io/pypa/manylinux_2_28_${ARCH}
|
||||||
|
|
||||||
ARG ARCH=x86_64
|
ARG ARCH=x86_64
|
||||||
ARG DOCKER_USER=default_user
|
ARG DOCKER_USER=default_user
|
||||||
|
|||||||
0
ci/manylinux_nodejs/build.sh → ci/manylinux_node/build_lancedb.sh
Executable file → Normal file
0
ci/manylinux_nodejs/build.sh → ci/manylinux_node/build_lancedb.sh
Executable file → Normal file
@@ -6,7 +6,7 @@
|
|||||||
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
||||||
set -e
|
set -e
|
||||||
|
|
||||||
git clone -b OpenSSL_1_1_1u \
|
git clone -b OpenSSL_1_1_1v \
|
||||||
--single-branch \
|
--single-branch \
|
||||||
https://github.com/openssl/openssl.git
|
https://github.com/openssl/openssl.git
|
||||||
|
|
||||||
|
|||||||
@@ -8,7 +8,7 @@ install_node() {
|
|||||||
|
|
||||||
source "$HOME"/.bashrc
|
source "$HOME"/.bashrc
|
||||||
|
|
||||||
nvm install --no-progress 16
|
nvm install --no-progress 18
|
||||||
}
|
}
|
||||||
|
|
||||||
install_rust() {
|
install_rust() {
|
||||||
|
|||||||
@@ -1,31 +0,0 @@
|
|||||||
# Many linux dockerfile with Rust, Node, and Lance dependencies installed.
|
|
||||||
# This container allows building the node modules native libraries in an
|
|
||||||
# environment with a very old glibc, so that we are compatible with a wide
|
|
||||||
# range of linux distributions.
|
|
||||||
ARG ARCH=x86_64
|
|
||||||
|
|
||||||
FROM quay.io/pypa/manylinux2014_${ARCH}
|
|
||||||
|
|
||||||
ARG ARCH=x86_64
|
|
||||||
ARG DOCKER_USER=default_user
|
|
||||||
|
|
||||||
# Install static openssl
|
|
||||||
COPY install_openssl.sh install_openssl.sh
|
|
||||||
RUN ./install_openssl.sh ${ARCH} > /dev/null
|
|
||||||
|
|
||||||
# Protobuf is also installed as root.
|
|
||||||
COPY install_protobuf.sh install_protobuf.sh
|
|
||||||
RUN ./install_protobuf.sh ${ARCH}
|
|
||||||
|
|
||||||
ENV DOCKER_USER=${DOCKER_USER}
|
|
||||||
# Create a group and user
|
|
||||||
RUN echo ${ARCH} && adduser --user-group --create-home --uid ${DOCKER_USER} build_user
|
|
||||||
|
|
||||||
# We switch to the user to install Rust and Node, since those like to be
|
|
||||||
# installed at the user level.
|
|
||||||
USER ${DOCKER_USER}
|
|
||||||
|
|
||||||
COPY prepare_manylinux_node.sh prepare_manylinux_node.sh
|
|
||||||
RUN cp /prepare_manylinux_node.sh $HOME/ && \
|
|
||||||
cd $HOME && \
|
|
||||||
./prepare_manylinux_node.sh ${ARCH}
|
|
||||||
@@ -1,26 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
# Builds openssl from source so we can statically link to it
|
|
||||||
|
|
||||||
# this is to avoid the error we get with the system installation:
|
|
||||||
# /usr/bin/ld: <library>: version node not found for symbol SSLeay@@OPENSSL_1.0.1
|
|
||||||
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
|
||||||
set -e
|
|
||||||
|
|
||||||
git clone -b OpenSSL_1_1_1u \
|
|
||||||
--single-branch \
|
|
||||||
https://github.com/openssl/openssl.git
|
|
||||||
|
|
||||||
pushd openssl
|
|
||||||
|
|
||||||
if [[ $1 == x86_64* ]]; then
|
|
||||||
ARCH=linux-x86_64
|
|
||||||
else
|
|
||||||
# gnu target
|
|
||||||
ARCH=linux-aarch64
|
|
||||||
fi
|
|
||||||
|
|
||||||
./Configure no-shared $ARCH
|
|
||||||
|
|
||||||
make
|
|
||||||
|
|
||||||
make install
|
|
||||||
@@ -1,15 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
# Installs protobuf compiler. Should be run as root.
|
|
||||||
set -e
|
|
||||||
|
|
||||||
if [[ $1 == x86_64* ]]; then
|
|
||||||
ARCH=x86_64
|
|
||||||
else
|
|
||||||
# gnu target
|
|
||||||
ARCH=aarch_64
|
|
||||||
fi
|
|
||||||
|
|
||||||
PB_REL=https://github.com/protocolbuffers/protobuf/releases
|
|
||||||
PB_VERSION=23.1
|
|
||||||
curl -LO $PB_REL/download/v$PB_VERSION/protoc-$PB_VERSION-linux-$ARCH.zip
|
|
||||||
unzip protoc-$PB_VERSION-linux-$ARCH.zip -d /usr/local
|
|
||||||
@@ -1,21 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
set -e
|
|
||||||
|
|
||||||
install_node() {
|
|
||||||
echo "Installing node..."
|
|
||||||
|
|
||||||
curl -o- https://raw.githubusercontent.com/nvm-sh/nvm/v0.34.0/install.sh | bash
|
|
||||||
|
|
||||||
source "$HOME"/.bashrc
|
|
||||||
|
|
||||||
nvm install --no-progress 16
|
|
||||||
}
|
|
||||||
|
|
||||||
install_rust() {
|
|
||||||
echo "Installing rust..."
|
|
||||||
curl https://sh.rustup.rs -sSf | bash -s -- -y
|
|
||||||
export PATH="$PATH:/root/.cargo/bin"
|
|
||||||
}
|
|
||||||
|
|
||||||
install_node
|
|
||||||
install_rust
|
|
||||||
@@ -100,6 +100,7 @@ nav:
|
|||||||
- Quickstart: reranking/index.md
|
- Quickstart: reranking/index.md
|
||||||
- Cohere Reranker: reranking/cohere.md
|
- Cohere Reranker: reranking/cohere.md
|
||||||
- Linear Combination Reranker: reranking/linear_combination.md
|
- Linear Combination Reranker: reranking/linear_combination.md
|
||||||
|
- Reciprocal Rank Fusion Reranker: reranking/rrf.md
|
||||||
- Cross Encoder Reranker: reranking/cross_encoder.md
|
- Cross Encoder Reranker: reranking/cross_encoder.md
|
||||||
- ColBERT Reranker: reranking/colbert.md
|
- ColBERT Reranker: reranking/colbert.md
|
||||||
- Jina Reranker: reranking/jina.md
|
- Jina Reranker: reranking/jina.md
|
||||||
@@ -140,10 +141,13 @@ nav:
|
|||||||
- Overview: examples/index.md
|
- Overview: examples/index.md
|
||||||
- 🐍 Python:
|
- 🐍 Python:
|
||||||
- Overview: examples/examples_python.md
|
- Overview: examples/examples_python.md
|
||||||
|
- Build From Scratch: examples/python_examples/build_from_scratch.md
|
||||||
|
- Multimodal: examples/python_examples/multimodal.md
|
||||||
|
- Rag: examples/python_examples/rag.md
|
||||||
|
- Miscellaneous:
|
||||||
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
||||||
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
||||||
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
||||||
- Example - Calculate CLIP Embeddings with Roboflow Inference: examples/image_embeddings_roboflow.md
|
|
||||||
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
||||||
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
||||||
- 👾 JavaScript:
|
- 👾 JavaScript:
|
||||||
@@ -185,6 +189,7 @@ nav:
|
|||||||
- Quickstart: reranking/index.md
|
- Quickstart: reranking/index.md
|
||||||
- Cohere Reranker: reranking/cohere.md
|
- Cohere Reranker: reranking/cohere.md
|
||||||
- Linear Combination Reranker: reranking/linear_combination.md
|
- Linear Combination Reranker: reranking/linear_combination.md
|
||||||
|
- Reciprocal Rank Fusion Reranker: reranking/rrf.md
|
||||||
- Cross Encoder Reranker: reranking/cross_encoder.md
|
- Cross Encoder Reranker: reranking/cross_encoder.md
|
||||||
- ColBERT Reranker: reranking/colbert.md
|
- ColBERT Reranker: reranking/colbert.md
|
||||||
- Jina Reranker: reranking/jina.md
|
- Jina Reranker: reranking/jina.md
|
||||||
@@ -219,14 +224,24 @@ nav:
|
|||||||
- PromptTools: integrations/prompttools.md
|
- PromptTools: integrations/prompttools.md
|
||||||
- Examples:
|
- Examples:
|
||||||
- examples/index.md
|
- examples/index.md
|
||||||
|
- 🐍 Python:
|
||||||
|
- Overview: examples/examples_python.md
|
||||||
|
- Build From Scratch: examples/python_examples/build_from_scratch.md
|
||||||
|
- Multimodal: examples/python_examples/multimodal.md
|
||||||
|
- Rag: examples/python_examples/rag.md
|
||||||
|
- Miscellaneous:
|
||||||
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
||||||
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
||||||
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
||||||
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
||||||
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
||||||
- YouTube Transcript Search (JS): examples/youtube_transcript_bot_with_nodejs.md
|
- 👾 JavaScript:
|
||||||
- Serverless Chatbot from any website: examples/serverless_website_chatbot.md
|
- Overview: examples/examples_js.md
|
||||||
|
- Serverless Website Chatbot: examples/serverless_website_chatbot.md
|
||||||
|
- YouTube Transcript Search: examples/youtube_transcript_bot_with_nodejs.md
|
||||||
- TransformersJS Embedding Search: examples/transformerjs_embedding_search_nodejs.md
|
- TransformersJS Embedding Search: examples/transformerjs_embedding_search_nodejs.md
|
||||||
|
- 🦀 Rust:
|
||||||
|
- Overview: examples/examples_rust.md
|
||||||
- API reference:
|
- API reference:
|
||||||
- Overview: api_reference.md
|
- Overview: api_reference.md
|
||||||
- Python: python/python.md
|
- Python: python/python.md
|
||||||
|
|||||||
1
docs/src/assets/colab.svg
Normal file
1
docs/src/assets/colab.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="117" height="20"><linearGradient id="b" x2="0" y2="100%"><stop offset="0" stop-color="#bbb" stop-opacity=".1"/><stop offset="1" stop-opacity=".1"/></linearGradient><clipPath id="a"><rect width="117" height="20" rx="3" fill="#fff"/></clipPath><g clip-path="url(#a)"><path fill="#555" d="M0 0h30v20H0z"/><path fill="#007ec6" d="M30 0h87v20H30z"/><path fill="url(#b)" d="M0 0h117v20H0z"/></g><g fill="#fff" text-anchor="middle" font-family="DejaVu Sans,Verdana,Geneva,sans-serif" font-size="110"><svg x="4px" y="0px" width="22px" height="20px" viewBox="-2 0 28 24" style="background-color: #fff;border-radius: 1px;"><path style="fill:#e8710a;" d="M1.977,16.77c-2.667-2.277-2.605-7.079,0-9.357C2.919,8.057,3.522,9.075,4.49,9.691c-1.152,1.6-1.146,3.201-0.004,4.803C3.522,15.111,2.918,16.126,1.977,16.77z"/><path style="fill:#f9ab00;" d="M12.257,17.114c-1.767-1.633-2.485-3.658-2.118-6.02c0.451-2.91,2.139-4.893,4.946-5.678c2.565-0.718,4.964-0.217,6.878,1.819c-0.884,0.743-1.707,1.547-2.434,2.446C18.488,8.827,17.319,8.435,16,8.856c-2.404,0.767-3.046,3.241-1.494,5.644c-0.241,0.275-0.493,0.541-0.721,0.826C13.295,15.939,12.511,16.3,12.257,17.114z"/><path style="fill:#e8710a;" d="M19.529,9.682c0.727-0.899,1.55-1.703,2.434-2.446c2.703,2.783,2.701,7.031-0.005,9.764c-2.648,2.674-6.936,2.725-9.701,0.115c0.254-0.814,1.038-1.175,1.528-1.788c0.228-0.285,0.48-0.552,0.721-0.826c1.053,0.916,2.254,1.268,3.6,0.83C20.502,14.551,21.151,11.927,19.529,9.682z"/><path style="fill:#f9ab00;" d="M4.49,9.691C3.522,9.075,2.919,8.057,1.977,7.413c2.209-2.398,5.721-2.942,8.476-1.355c0.555,0.32,0.719,0.606,0.285,1.128c-0.157,0.188-0.258,0.422-0.391,0.631c-0.299,0.47-0.509,1.067-0.929,1.371C8.933,9.539,8.523,8.847,8.021,8.746C6.673,8.475,5.509,8.787,4.49,9.691z"/><path style="fill:#f9ab00;" d="M1.977,16.77c0.941-0.644,1.545-1.659,2.509-2.277c1.373,1.152,2.85,1.433,4.45,0.499c0.332-0.194,0.503-0.088,0.673,0.19c0.386,0.635,0.753,1.285,1.181,1.89c0.34,0.48,0.222,0.715-0.253,1.006C7.84,19.73,4.205,19.188,1.977,16.77z"/></svg><text x="245" y="140" transform="scale(.1)" textLength="30"> </text><text x="725" y="150" fill="#010101" fill-opacity=".3" transform="scale(.1)" textLength="770">Open in Colab</text><text x="725" y="140" transform="scale(.1)" textLength="770">Open in Colab</text></g> </svg>
|
||||||
|
After Width: | Height: | Size: 2.3 KiB |
1
docs/src/assets/ghost.svg
Normal file
1
docs/src/assets/ghost.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="88.25" height="28" role="img" aria-label="GHOST"><title>GHOST</title><g shape-rendering="crispEdges"><rect width="88.25" height="28" fill="#000"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSIjZjdkZjFlIiByb2xlPSJpbWciIHZpZXdCb3g9IjAgMCAyNCAyNCIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIj48dGl0bGU+R2hvc3Q8L3RpdGxlPjxwYXRoIGQ9Ik0xMiAwQzUuMzczIDAgMCA1LjM3MyAwIDEyczUuMzczIDEyIDEyIDEyIDEyLTUuMzczIDEyLTEyUzE4LjYyNyAwIDEyIDB6bS4yNTYgMi4zMTNjMi40Ny4wMDUgNS4xMTYgMi4wMDggNS44OTggMi45NjJsLjI0NC4zYzEuNjQgMS45OTQgMy41NjkgNC4zNCAzLjU2OSA2Ljk2NiAwIDMuNzE5LTIuOTggNS44MDgtNi4xNTggNy41MDgtMS40MzMuNzY2LTIuOTggMS41MDgtNC43NDggMS41MDgtNC41NDMgMC04LjM2Ni0zLjU2OS04LjM2Ni04LjExMiAwLS43MDYuMTctMS40MjUuMzQyLTIuMTUuMTIyLS41MTUuMjQ0LTEuMDMzLjMwNy0xLjU0OS41NDgtNC41MzkgMi45NjctNi43OTUgOC40MjItNy40MDhhNC4yOSA0LjI5IDAgMDEuNDktLjAyNloiLz48L3N2Zz4="/><text transform="scale(.1)" x="541.25" y="175" textLength="442.5" fill="#fff" font-weight="bold">GHOST</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 1.2 KiB |
1
docs/src/assets/github.svg
Normal file
1
docs/src/assets/github.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="95.5" height="28" role="img" aria-label="GITHUB"><title>GITHUB</title><g shape-rendering="crispEdges"><rect width="95.5" height="28" fill="#121011"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSJ3aGl0ZSIgcm9sZT0iaW1nIiB2aWV3Qm94PSIwIDAgMjQgMjQiIHhtbG5zPSJodHRwOi8vd3d3LnczLm9yZy8yMDAwL3N2ZyI+PHRpdGxlPkdpdEh1YjwvdGl0bGU+PHBhdGggZD0iTTEyIC4yOTdjLTYuNjMgMC0xMiA1LjM3My0xMiAxMiAwIDUuMzAzIDMuNDM4IDkuOCA4LjIwNSAxMS4zODUuNi4xMTMuODItLjI1OC44Mi0uNTc3IDAtLjI4NS0uMDEtMS4wNC0uMDE1LTIuMDQtMy4zMzguNzI0LTQuMDQyLTEuNjEtNC4wNDItMS42MUM0LjQyMiAxOC4wNyAzLjYzMyAxNy43IDMuNjMzIDE3LjdjLTEuMDg3LS43NDQuMDg0LS43MjkuMDg0LS43MjkgMS4yMDUuMDg0IDEuODM4IDEuMjM2IDEuODM4IDEuMjM2IDEuMDcgMS44MzUgMi44MDkgMS4zMDUgMy40OTUuOTk4LjEwOC0uNzc2LjQxNy0xLjMwNS43Ni0xLjYwNS0yLjY2NS0uMy01LjQ2Ni0xLjMzMi01LjQ2Ni01LjkzIDAtMS4zMS40NjUtMi4zOCAxLjIzNS0zLjIyLS4xMzUtLjMwMy0uNTQtMS41MjMuMTA1LTMuMTc2IDAgMCAxLjAwNS0uMzIyIDMuMyAxLjIzLjk2LS4yNjcgMS45OC0uMzk5IDMtLjQwNSAxLjAyLjAwNiAyLjA0LjEzOCAzIC40MDUgMi4yOC0xLjU1MiAzLjI4NS0xLjIzIDMuMjg1LTEuMjMuNjQ1IDEuNjUzLjI0IDIuODczLjEyIDMuMTc2Ljc2NS44NCAxLjIzIDEuOTEgMS4yMyAzLjIyIDAgNC42MS0yLjgwNSA1LjYyNS01LjQ3NSA1LjkyLjQyLjM2LjgxIDEuMDk2LjgxIDIuMjIgMCAxLjYwNi0uMDE1IDIuODk2LS4wMTUgMy4yODYgMCAuMzE1LjIxLjY5LjgyNS41N0MyMC41NjUgMjIuMDkyIDI0IDE3LjU5MiAyNCAxMi4yOTdjMC02LjYyNy01LjM3My0xMi0xMi0xMiIvPjwvc3ZnPg=="/><text transform="scale(.1)" x="577.5" y="175" textLength="515" fill="#fff" font-weight="bold">GITHUB</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 1.7 KiB |
1
docs/src/assets/python.svg
Normal file
1
docs/src/assets/python.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="97.5" height="28" role="img" aria-label="PYTHON"><title>PYTHON</title><g shape-rendering="crispEdges"><rect width="97.5" height="28" fill="#3670a0"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSIjZmZkZDU0IiByb2xlPSJpbWciIHZpZXdCb3g9IjAgMCAyNCAyNCIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIj48dGl0bGU+UHl0aG9uPC90aXRsZT48cGF0aCBkPSJNMTQuMjUuMThsLjkuMi43My4yNi41OS4zLjQ1LjMyLjM0LjM0LjI1LjM0LjE2LjMzLjEuMy4wNC4yNi4wMi4yLS4wMS4xM1Y4LjVsLS4wNS42My0uMTMuNTUtLjIxLjQ2LS4yNi4zOC0uMy4zMS0uMzMuMjUtLjM1LjE5LS4zNS4xNC0uMzMuMS0uMy4wNy0uMjYuMDQtLjIxLjAySDguNzdsLS42OS4wNS0uNTkuMTQtLjUuMjItLjQxLjI3LS4zMy4zMi0uMjcuMzUtLjIuMzYtLjE1LjM3LS4xLjM1LS4wNy4zMi0uMDQuMjctLjAyLjIxdjMuMDZIMy4xN2wtLjIxLS4wMy0uMjgtLjA3LS4zMi0uMTItLjM1LS4xOC0uMzYtLjI2LS4zNi0uMzYtLjM1LS40Ni0uMzItLjU5LS4yOC0uNzMtLjIxLS44OC0uMTQtMS4wNS0uMDUtMS4yMy4wNi0xLjIyLjE2LTEuMDQuMjQtLjg3LjMyLS43MS4zNi0uNTcuNC0uNDQuNDItLjMzLjQyLS4yNC40LS4xNi4zNi0uMS4zMi0uMDUuMjQtLjAxaC4xNmwuMDYuMDFoOC4xNnYtLjgzSDYuMThsLS4wMS0yLjc1LS4wMi0uMzcuMDUtLjM0LjExLS4zMS4xNy0uMjguMjUtLjI2LjMxLS4yMy4zOC0uMi40NC0uMTguNTEtLjE1LjU4LS4xMi42NC0uMS43MS0uMDYuNzctLjA0Ljg0LS4wMiAxLjI3LjA1em0tNi4zIDEuOThsLS4yMy4zMy0uMDguNDEuMDguNDEuMjMuMzQuMzMuMjIuNDEuMDkuNDEtLjA5LjMzLS4yMi4yMy0uMzQuMDgtLjQxLS4wOC0uNDEtLjIzLS4zMy0uMzMtLjIyLS40MS0uMDktLjQxLjA5em0xMy4wOSAzLjk1bC4yOC4wNi4zMi4xMi4zNS4xOC4zNi4yNy4zNi4zNS4zNS40Ny4zMi41OS4yOC43My4yMS44OC4xNCAxLjA0LjA1IDEuMjMtLjA2IDEuMjMtLjE2IDEuMDQtLjI0Ljg2LS4zMi43MS0uMzYuNTctLjQuNDUtLjQyLjMzLS40Mi4yNC0uNC4xNi0uMzYuMDktLjMyLjA1LS4yNC4wMi0uMTYtLjAxaC04LjIydi44Mmg1Ljg0bC4wMSAyLjc2LjAyLjM2LS4wNS4zNC0uMTEuMzEtLjE3LjI5LS4yNS4yNS0uMzEuMjQtLjM4LjItLjQ0LjE3LS41MS4xNS0uNTguMTMtLjY0LjA5LS43MS4wNy0uNzcuMDQtLjg0LjAxLTEuMjctLjA0LTEuMDctLjE0LS45LS4yLS43My0uMjUtLjU5LS4zLS40NS0uMzMtLjM0LS4zNC0uMjUtLjM0LS4xNi0uMzMtLjEtLjMtLjA0LS4yNS0uMDItLjIuMDEtLjEzdi01LjM0bC4wNS0uNjQuMTMtLjU0LjIxLS40Ni4yNi0uMzguMy0uMzIuMzMtLjI0LjM1LS4yLjM1LS4xNC4zMy0uMS4zLS4wNi4yNi0uMDQuMjEtLjAyLjEzLS4wMWg1Ljg0bC42OS0uMDUuNTktLjE0LjUtLjIxLjQxLS4yOC4zMy0uMzIuMjctLjM1LjItLjM2LjE1LS4zNi4xLS4zNS4wNy0uMzIuMDQtLjI4LjAyLS4yMVY2LjA3aDIuMDlsLjE0LjAxem0tNi40NyAxNC4yNWwtLjIzLjMzLS4wOC40MS4wOC40MS4yMy4zMy4zMy4yMy40MS4wOC40MS0uMDguMzMtLjIzLjIzLS4zMy4wOC0uNDEtLjA4LS40MS0uMjMtLjMzLS4zMy0uMjMtLjQxLS4wOC0uNDEuMDh6Ii8+PC9zdmc+"/><text transform="scale(.1)" x="587.5" y="175" textLength="535" fill="#fff" font-weight="bold">PYTHON</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 2.6 KiB |
@@ -15,12 +15,15 @@ There is another optional layer of abstraction available: `TextEmbeddingFunction
|
|||||||
|
|
||||||
Let's implement `SentenceTransformerEmbeddings` class. All you need to do is implement the `generate_embeddings()` and `ndims` function to handle the input types you expect and register the class in the global `EmbeddingFunctionRegistry`
|
Let's implement `SentenceTransformerEmbeddings` class. All you need to do is implement the `generate_embeddings()` and `ndims` function to handle the input types you expect and register the class in the global `EmbeddingFunctionRegistry`
|
||||||
|
|
||||||
```python
|
|
||||||
from lancedb.embeddings import register
|
|
||||||
from lancedb.util import attempt_import_or_raise
|
|
||||||
|
|
||||||
@register("sentence-transformers")
|
=== "Python"
|
||||||
class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
|
||||||
|
```python
|
||||||
|
from lancedb.embeddings import register
|
||||||
|
from lancedb.util import attempt_import_or_raise
|
||||||
|
|
||||||
|
@register("sentence-transformers")
|
||||||
|
class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
||||||
name: str = "all-MiniLM-L6-v2"
|
name: str = "all-MiniLM-L6-v2"
|
||||||
# set more default instance vars like device, etc.
|
# set more default instance vars like device, etc.
|
||||||
|
|
||||||
@@ -39,38 +42,59 @@ class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
|||||||
@cached(cache={})
|
@cached(cache={})
|
||||||
def _embedding_model(self):
|
def _embedding_model(self):
|
||||||
return sentence_transformers.SentenceTransformer(name)
|
return sentence_transformers.SentenceTransformer(name)
|
||||||
```
|
```
|
||||||
|
|
||||||
This is a stripped down version of our implementation of `SentenceTransformerEmbeddings` that removes certain optimizations and defaul settings.
|
=== "TypeScript"
|
||||||
|
|
||||||
|
```ts
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:imports"
|
||||||
|
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:embedding_impl"
|
||||||
|
```
|
||||||
|
|
||||||
|
|
||||||
|
This is a stripped down version of our implementation of `SentenceTransformerEmbeddings` that removes certain optimizations and default settings.
|
||||||
|
|
||||||
Now you can use this embedding function to create your table schema and that's it! you can then ingest data and run queries without manually vectorizing the inputs.
|
Now you can use this embedding function to create your table schema and that's it! you can then ingest data and run queries without manually vectorizing the inputs.
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
from lancedb.pydantic import LanceModel, Vector
|
|
||||||
|
|
||||||
registry = EmbeddingFunctionRegistry.get_instance()
|
```python
|
||||||
stransformer = registry.get("sentence-transformers").create()
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
|
||||||
class TextModelSchema(LanceModel):
|
registry = EmbeddingFunctionRegistry.get_instance()
|
||||||
|
stransformer = registry.get("sentence-transformers").create()
|
||||||
|
|
||||||
|
class TextModelSchema(LanceModel):
|
||||||
vector: Vector(stransformer.ndims) = stransformer.VectorField()
|
vector: Vector(stransformer.ndims) = stransformer.VectorField()
|
||||||
text: str = stransformer.SourceField()
|
text: str = stransformer.SourceField()
|
||||||
|
|
||||||
tbl = db.create_table("table", schema=TextModelSchema)
|
tbl = db.create_table("table", schema=TextModelSchema)
|
||||||
|
|
||||||
tbl.add(pd.DataFrame({"text": ["halo", "world"]}))
|
tbl.add(pd.DataFrame({"text": ["halo", "world"]}))
|
||||||
result = tbl.search("world").limit(5)
|
result = tbl.search("world").limit(5)
|
||||||
```
|
```
|
||||||
|
|
||||||
NOTE:
|
=== "TypeScript"
|
||||||
|
|
||||||
You can always implement the `EmbeddingFunction` interface directly if you want or need to, `TextEmbeddingFunction` just makes it much simpler and faster for you to do so, by setting up the boiler plat for text-specific use case
|
```ts
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:call_custom_function"
|
||||||
|
```
|
||||||
|
|
||||||
|
!!! note
|
||||||
|
|
||||||
|
You can always implement the `EmbeddingFunction` interface directly if you want or need to, `TextEmbeddingFunction` just makes it much simpler and faster for you to do so, by setting up the boiler plat for text-specific use case
|
||||||
|
|
||||||
## Multi-modal embedding function example
|
## Multi-modal embedding function example
|
||||||
You can also use the `EmbeddingFunction` interface to implement more complex workflows such as multi-modal embedding function support. LanceDB implements `OpenClipEmeddingFunction` class that suppports multi-modal seach. Here's the implementation that you can use as a reference to build your own multi-modal embedding functions.
|
You can also use the `EmbeddingFunction` interface to implement more complex workflows such as multi-modal embedding function support.
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
@register("open-clip")
|
|
||||||
class OpenClipEmbeddings(EmbeddingFunction):
|
LanceDB implements `OpenClipEmeddingFunction` class that suppports multi-modal seach. Here's the implementation that you can use as a reference to build your own multi-modal embedding functions.
|
||||||
|
|
||||||
|
```python
|
||||||
|
@register("open-clip")
|
||||||
|
class OpenClipEmbeddings(EmbeddingFunction):
|
||||||
name: str = "ViT-B-32"
|
name: str = "ViT-B-32"
|
||||||
pretrained: str = "laion2b_s34b_b79k"
|
pretrained: str = "laion2b_s34b_b79k"
|
||||||
device: str = "cpu"
|
device: str = "cpu"
|
||||||
@@ -209,4 +233,8 @@ class OpenClipEmbeddings(EmbeddingFunction):
|
|||||||
if self.normalize:
|
if self.normalize:
|
||||||
image_features /= image_features.norm(dim=-1, keepdim=True)
|
image_features /= image_features.norm(dim=-1, keepdim=True)
|
||||||
return image_features.cpu().numpy().squeeze()
|
return image_features.cpu().numpy().squeeze()
|
||||||
```
|
```
|
||||||
|
|
||||||
|
=== "TypeScript"
|
||||||
|
|
||||||
|
Coming Soon! See this [issue](https://github.com/lancedb/lancedb/issues/1482) to track the status!
|
||||||
|
|||||||
@@ -390,6 +390,7 @@ Supported parameters (to be passed in `create` method) are:
|
|||||||
| `query_input_type` | `str` | `"search_query"` | The type of input data to be used for the query. |
|
| `query_input_type` | `str` | `"search_query"` | The type of input data to be used for the query. |
|
||||||
|
|
||||||
Cohere supports following input types:
|
Cohere supports following input types:
|
||||||
|
|
||||||
| Input Type | Description |
|
| Input Type | Description |
|
||||||
|-------------------------|---------------------------------------|
|
|-------------------------|---------------------------------------|
|
||||||
| "`search_document`" | Used for embeddings stored in a vector|
|
| "`search_document`" | Used for embeddings stored in a vector|
|
||||||
@@ -517,6 +518,82 @@ tbl.add(df)
|
|||||||
rs = tbl.search("hello").limit(1).to_pandas()
|
rs = tbl.search("hello").limit(1).to_pandas()
|
||||||
```
|
```
|
||||||
|
|
||||||
|
# IBM watsonx.ai Embeddings
|
||||||
|
|
||||||
|
Generate text embeddings using IBM's watsonx.ai platform.
|
||||||
|
|
||||||
|
## Supported Models
|
||||||
|
|
||||||
|
You can find a list of supported models at [IBM watsonx.ai Documentation](https://dataplatform.cloud.ibm.com/docs/content/wsj/analyze-data/fm-models-embed.html?context=wx). The currently supported model names are:
|
||||||
|
|
||||||
|
- `ibm/slate-125m-english-rtrvr`
|
||||||
|
- `ibm/slate-30m-english-rtrvr`
|
||||||
|
- `sentence-transformers/all-minilm-l12-v2`
|
||||||
|
- `intfloat/multilingual-e5-large`
|
||||||
|
|
||||||
|
## Parameters
|
||||||
|
|
||||||
|
The following parameters can be passed to the `create` method:
|
||||||
|
|
||||||
|
| Parameter | Type | Default Value | Description |
|
||||||
|
|------------|----------|----------------------------------|-----------------------------------------------------------|
|
||||||
|
| name | str | "ibm/slate-125m-english-rtrvr" | The model ID of the watsonx.ai model to use |
|
||||||
|
| api_key | str | None | Optional IBM Cloud API key (or set `WATSONX_API_KEY`) |
|
||||||
|
| project_id | str | None | Optional watsonx project ID (or set `WATSONX_PROJECT_ID`) |
|
||||||
|
| url | str | None | Optional custom URL for the watsonx.ai instance |
|
||||||
|
| params | dict | None | Optional additional parameters for the embedding model |
|
||||||
|
|
||||||
|
## Usage Example
|
||||||
|
|
||||||
|
First, the watsonx.ai library is an optional dependency, so must be installed seperately:
|
||||||
|
|
||||||
|
```
|
||||||
|
pip install ibm-watsonx-ai
|
||||||
|
```
|
||||||
|
|
||||||
|
Optionally set environment variables (if not passing credentials to `create` directly):
|
||||||
|
|
||||||
|
```sh
|
||||||
|
export WATSONX_API_KEY="YOUR_WATSONX_API_KEY"
|
||||||
|
export WATSONX_PROJECT_ID="YOUR_WATSONX_PROJECT_ID"
|
||||||
|
```
|
||||||
|
|
||||||
|
```python
|
||||||
|
import os
|
||||||
|
import lancedb
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.embeddings import EmbeddingFunctionRegistry
|
||||||
|
|
||||||
|
watsonx_embed = EmbeddingFunctionRegistry
|
||||||
|
.get_instance()
|
||||||
|
.get("watsonx")
|
||||||
|
.create(
|
||||||
|
name="ibm/slate-125m-english-rtrvr",
|
||||||
|
# Uncomment and set these if not using environment variables
|
||||||
|
# api_key="your_api_key_here",
|
||||||
|
# project_id="your_project_id_here",
|
||||||
|
# url="your_watsonx_url_here",
|
||||||
|
# params={...},
|
||||||
|
)
|
||||||
|
|
||||||
|
class TextModel(LanceModel):
|
||||||
|
text: str = watsonx_embed.SourceField()
|
||||||
|
vector: Vector(watsonx_embed.ndims()) = watsonx_embed.VectorField()
|
||||||
|
|
||||||
|
data = [
|
||||||
|
{"text": "hello world"},
|
||||||
|
{"text": "goodbye world"},
|
||||||
|
]
|
||||||
|
|
||||||
|
db = lancedb.connect("~/.lancedb")
|
||||||
|
tbl = db.create_table("watsonx_test", schema=TextModel, mode="overwrite")
|
||||||
|
|
||||||
|
tbl.add(data)
|
||||||
|
|
||||||
|
rs = tbl.search("hello").limit(1).to_pandas()
|
||||||
|
print(rs)
|
||||||
|
```
|
||||||
|
|
||||||
## Multi-modal embedding functions
|
## Multi-modal embedding functions
|
||||||
Multi-modal embedding functions allow you to query your table using both images and text.
|
Multi-modal embedding functions allow you to query your table using both images and text.
|
||||||
|
|
||||||
|
|||||||
@@ -2,8 +2,8 @@ Representing multi-modal data as vector embeddings is becoming a standard practi
|
|||||||
|
|
||||||
For this purpose, LanceDB introduces an **embedding functions API**, that allow you simply set up once, during the configuration stage of your project. After this, the table remembers it, effectively making the embedding functions *disappear in the background* so you don't have to worry about manually passing callables, and instead, simply focus on the rest of your data engineering pipeline.
|
For this purpose, LanceDB introduces an **embedding functions API**, that allow you simply set up once, during the configuration stage of your project. After this, the table remembers it, effectively making the embedding functions *disappear in the background* so you don't have to worry about manually passing callables, and instead, simply focus on the rest of your data engineering pipeline.
|
||||||
|
|
||||||
!!! Note "LanceDB cloud doesn't support embedding functions yet"
|
!!! Note "Embedding functions on LanceDB cloud"
|
||||||
LanceDB Cloud does not support embedding functions yet. You need to generate embeddings before ingesting into the table or querying.
|
When using embedding functions with LanceDB cloud, the embeddings will be generated on the source device and sent to the cloud. This means that the source device must have the necessary resources to generate the embeddings.
|
||||||
|
|
||||||
!!! warning
|
!!! warning
|
||||||
Using the embedding function registry means that you don't have to explicitly generate the embeddings yourself.
|
Using the embedding function registry means that you don't have to explicitly generate the embeddings yourself.
|
||||||
|
|||||||
@@ -99,28 +99,28 @@ LanceDB registers the Sentence Transformers embeddings function in the registry
|
|||||||
|
|
||||||
Coming Soon!
|
Coming Soon!
|
||||||
|
|
||||||
### Jina Embeddings
|
### Embedding function with LanceDB cloud
|
||||||
|
Embedding functions are now supported on LanceDB cloud. The embeddings will be generated on the source device and sent to the cloud. This means that the source device must have the necessary resources to generate the embeddings. Here's an example using the OpenAI embedding function:
|
||||||
LanceDB registers the JinaAI embeddings function in the registry as `jina`. You can pass any supported model name to the `create`. By default it uses `"jina-clip-v1"`.
|
|
||||||
`jina-clip-v1` can handle both text and images and other models only support `text`.
|
|
||||||
|
|
||||||
You need to pass `JINA_API_KEY` in the environment variable or pass it as `api_key` to `create` method.
|
|
||||||
|
|
||||||
```python
|
```python
|
||||||
import os
|
import os
|
||||||
import lancedb
|
import lancedb
|
||||||
from lancedb.pydantic import LanceModel, Vector
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
from lancedb.embeddings import get_registry
|
from lancedb.embeddings import get_registry
|
||||||
os.environ['JINA_API_KEY'] = "jina_*"
|
os.environ['OPENAI_API_KEY'] = "..."
|
||||||
|
|
||||||
db = lancedb.connect("/tmp/db")
|
db = lancedb.connect(
|
||||||
func = get_registry().get("jina").create(name="jina-clip-v1")
|
uri="db://....",
|
||||||
|
api_key="sk_...",
|
||||||
|
region="us-east-1"
|
||||||
|
)
|
||||||
|
func = get_registry().get("openai").create()
|
||||||
|
|
||||||
class Words(LanceModel):
|
class Words(LanceModel):
|
||||||
text: str = func.SourceField()
|
text: str = func.SourceField()
|
||||||
vector: Vector(func.ndims()) = func.VectorField()
|
vector: Vector(func.ndims()) = func.VectorField()
|
||||||
|
|
||||||
table = db.create_table("words", schema=Words, mode="overwrite")
|
table = db.create_table("words", schema=Words)
|
||||||
table.add(
|
table.add(
|
||||||
[
|
[
|
||||||
{"text": "hello world"},
|
{"text": "hello world"},
|
||||||
|
|||||||
@@ -10,7 +10,7 @@ LanceDB provides language APIs, allowing you to embed a database in your languag
|
|||||||
|
|
||||||
## Applications powered by LanceDB
|
## Applications powered by LanceDB
|
||||||
|
|
||||||
| Project Name | Description | Screenshot |
|
| Project Name | Description |
|
||||||
|-----------------------------------------------------|----------------------------------------------------------------------------------------------------------------------|-------------------------------------------|
|
| --- | --- |
|
||||||
| [YOLOExplorer](https://github.com/lancedb/yoloexplorer) | Iterate on your YOLO / CV datasets using SQL, Vector semantic search, and more within seconds | 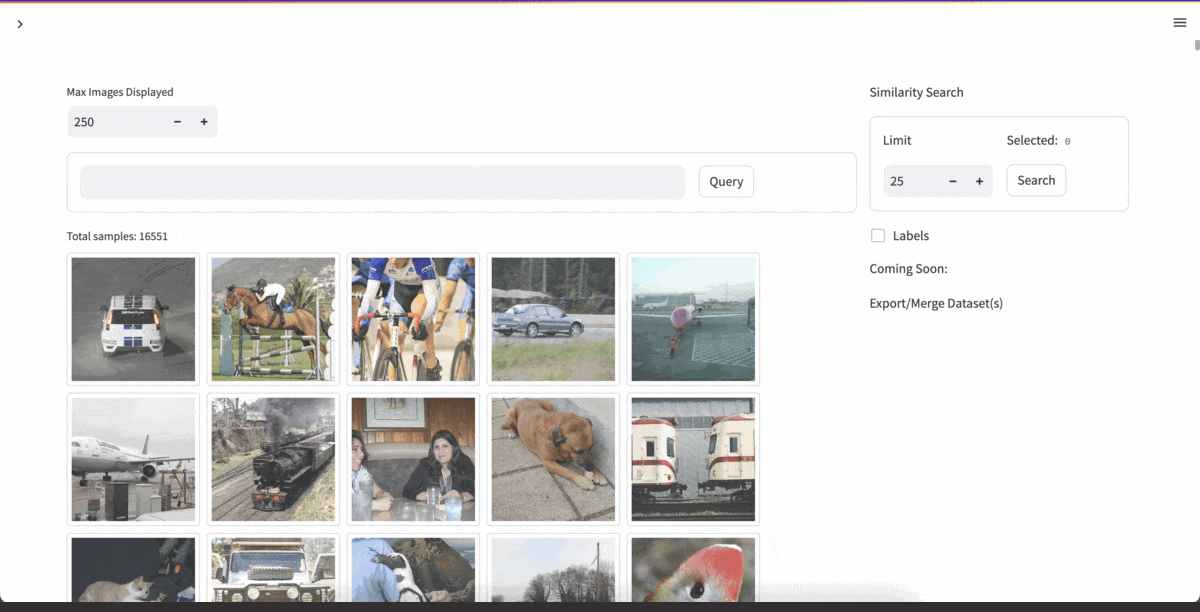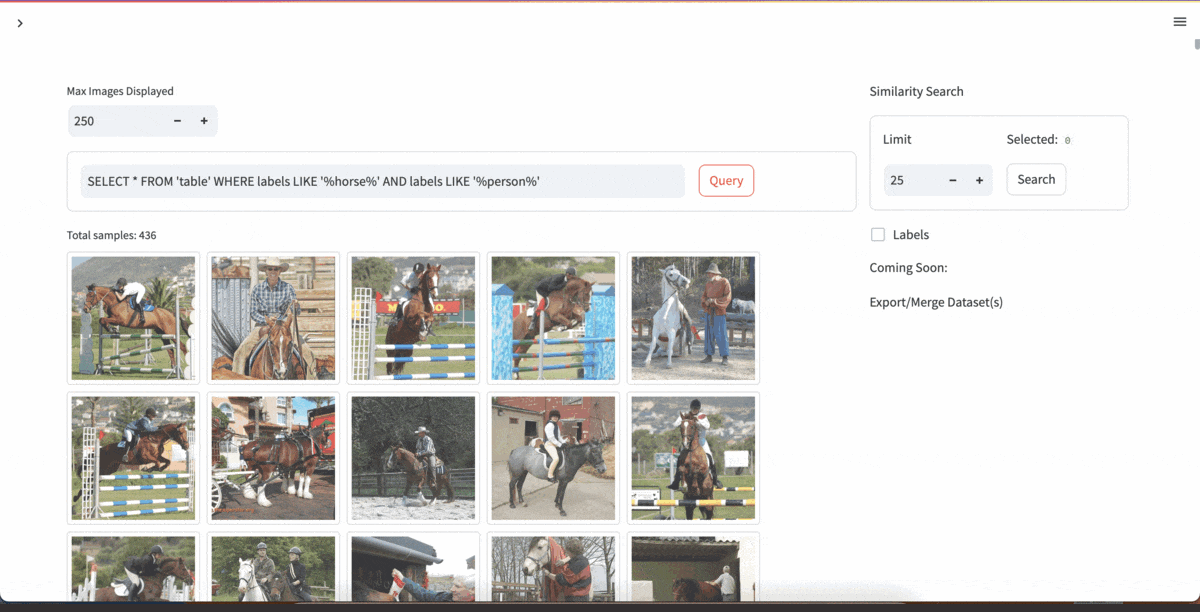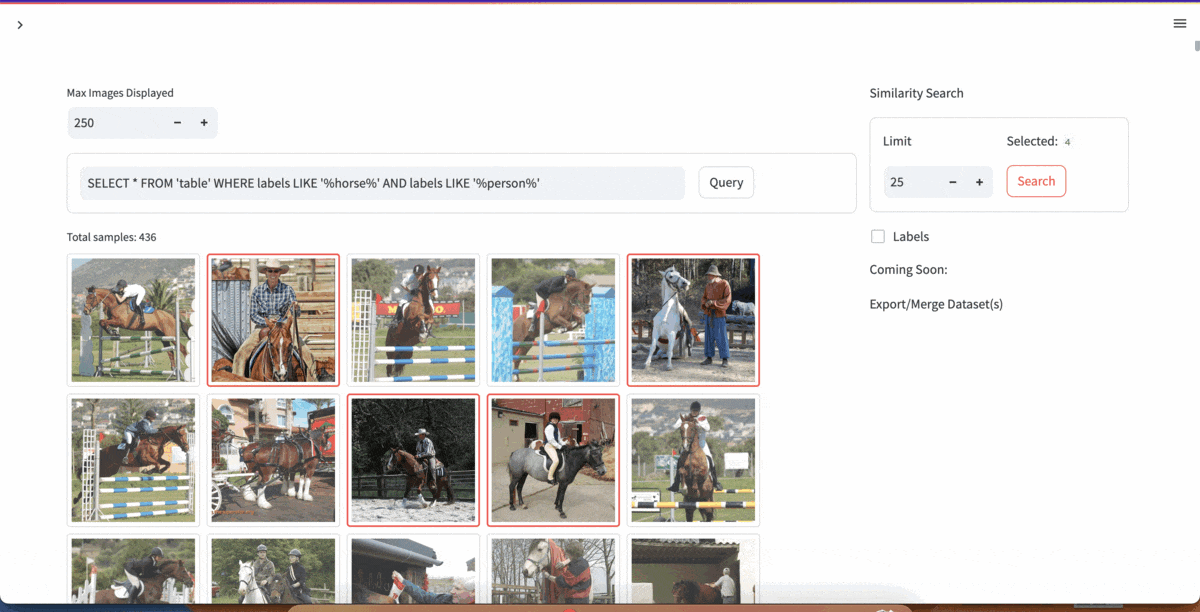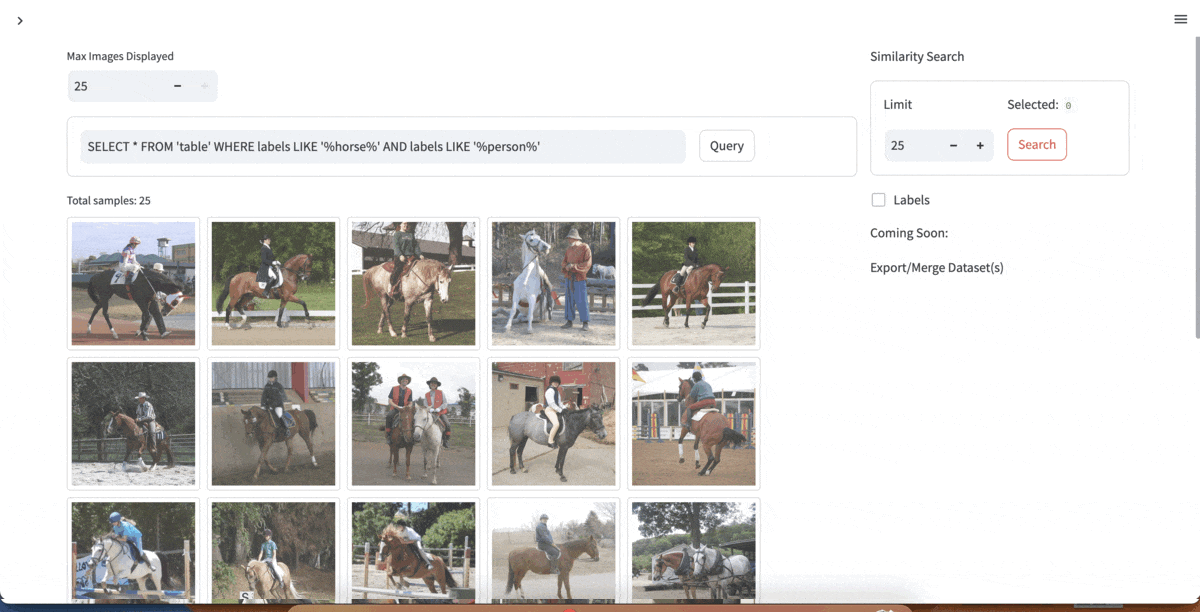 |
|
| **Ultralytics Explorer 🚀**<br>[](https://docs.ultralytics.com/datasets/explorer/)<br>[](https://colab.research.google.com/github/ultralytics/ultralytics/blob/main/docs/en/datasets/explorer/explorer.ipynb) | - 🔍 **Explore CV Datasets**: Semantic search, SQL queries, vector similarity, natural language.<br>- 🖥️ **GUI & Python API**: Seamless dataset interaction.<br>- ⚡ **Efficient & Scalable**: Leverages LanceDB for large datasets.<br>- 📊 **Detailed Analysis**: Easily analyze data patterns.<br>- 🌐 **Browser GUI Demo**: Create embeddings, search images, run queries. |
|
||||||
| [Website Chatbot (Deployable Vercel Template)](https://github.com/lancedb/lancedb-vercel-chatbot) | Create a chatbot from the sitemap of any website/docs of your choice. Built using vectorDB serverless native javascript package. |  |
|
| **Website Chatbot🤖**<br>[](https://github.com/lancedb/lancedb-vercel-chatbot)<br>[](https://vercel.com/new/clone?repository-url=https%3A%2F%2Fgithub.com%2Flancedb%2Flancedb-vercel-chatbot&env=OPENAI_API_KEY&envDescription=OpenAI%20API%20Key%20for%20chat%20completion.&project-name=lancedb-vercel-chatbot&repository-name=lancedb-vercel-chatbot&demo-title=LanceDB%20Chatbot%20Demo&demo-description=Demo%20website%20chatbot%20with%20LanceDB.&demo-url=https%3A%2F%2Flancedb.vercel.app&demo-image=https%3A%2F%2Fi.imgur.com%2FazVJtvr.png) | - 🌐 **Chatbot from Sitemap/Docs**: Create a chatbot using site or document context.<br>- 🚀 **Embed LanceDB in Next.js**: Lightweight, on-prem storage.<br>- 🧠 **AI-Powered Context Retrieval**: Efficiently access relevant data.<br>- 🔧 **Serverless & Native JS**: Seamless integration with Next.js.<br>- ⚡ **One-Click Deploy on Vercel**: Quick and easy setup.. |
|
||||||
|
|||||||
13
docs/src/examples/python_examples/build_from_scratch.md
Normal file
13
docs/src/examples/python_examples/build_from_scratch.md
Normal file
@@ -0,0 +1,13 @@
|
|||||||
|
# Build from Scratch with LanceDB 🚀
|
||||||
|
|
||||||
|
Start building your GenAI applications from the ground up using LanceDB's efficient vector-based document retrieval capabilities! 📄
|
||||||
|
|
||||||
|
#### Get Started in Minutes ⏱️
|
||||||
|
|
||||||
|
These examples provide a solid foundation for building your own GenAI applications using LanceDB. Jump from idea to proof of concept quickly with applied examples. Get started and see what you can create! 💻
|
||||||
|
|
||||||
|
| **Build From Scratch** | **Description** | **Links** |
|
||||||
|
|:-------------------------------------------|:-------------------------------------------------------------------------------------------------------------|:-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
|
||||||
|
| **Build RAG from Scratch🚀💻** | 📝 Create a **Retrieval-Augmented Generation** (RAG) model from scratch using LanceDB. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/RAG-from-Scratch)<br>[]() |
|
||||||
|
| **Local RAG from Scratch with Llama3🔥💡** | 🐫 Build a local RAG model using **Llama3** and **LanceDB** for fast and efficient text generation. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Local-RAG-from-Scratch)<br>[](https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Local-RAG-from-Scratch/rag.py) |
|
||||||
|
| **Multi-Head RAG from Scratch📚💻** | 🤯 Develop a **Multi-Head RAG model** from scratch, enabling generation of text based on multiple documents. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Multi-Head-RAG-from-Scratch)<br>[](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Multi-Head-RAG-from-Scratch) |
|
||||||
28
docs/src/examples/python_examples/multimodal.md
Normal file
28
docs/src/examples/python_examples/multimodal.md
Normal file
@@ -0,0 +1,28 @@
|
|||||||
|
# Multimodal Search with LanceDB 🔍💡
|
||||||
|
|
||||||
|
Experience the future of search with LanceDB's multimodal capabilities. Combine text and image queries to find the most relevant results in your corpus and unlock new possibilities! 🔓💡
|
||||||
|
|
||||||
|
#### Explore the Future of Search 🚀
|
||||||
|
|
||||||
|
Unlock the power of multimodal search with LanceDB, enabling efficient vector-based retrieval of text and image data! 📊💻
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
| **Multimodal** | **Description** | **Links** |
|
||||||
|
|:----------------|:-----------------|:-----------|
|
||||||
|
| **Multimodal CLIP: DiffusionDB 🌐💥** | Revolutionize search with Multimodal CLIP and DiffusionDB, combining text and image understanding for a new dimension of discovery! 🔓 | [][Clip_diffusionDB_github] <br>[][Clip_diffusionDB_colab] <br>[][Clip_diffusionDB_python] <br>[][Clip_diffusionDB_ghost] |
|
||||||
|
| **Multimodal CLIP: Youtube Videos 📹👀** | Search Youtube videos using Multimodal CLIP, finding relevant content with ease and accuracy! 🎯 | [][Clip_youtube_github] <br>[][Clip_youtube_colab] <br> [][Clip_youtube_python] <br>[][Clip_youtube_python] |
|
||||||
|
| **Multimodal Image + Text Search 📸🔍** | Discover relevant documents and images with a single query, using LanceDB's multimodal search capabilities to bridge the gap between text and visuals! 🌉 | [](https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_search) <br>[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_search/main.ipynb) <br> [](https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_search/main.py)<br> [](https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/) |
|
||||||
|
| **Cambrian-1: Vision-Centric Image Exploration 🔍👀** | Dive into vision-centric exploration of images with Cambrian-1, powered by LanceDB's multimodal search to uncover new insights! 🔎 | [](https://www.kaggle.com/code/prasantdixit/cambrian-1-vision-centric-exploration-of-images/)<br>[]() <br> []() <br> [](https://blog.lancedb.com/cambrian-1-vision-centric-exploration/) |
|
||||||
|
|
||||||
|
|
||||||
|
[Clip_diffusionDB_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb
|
||||||
|
[Clip_diffusionDB_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb/main.ipynb
|
||||||
|
[Clip_diffusionDB_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb/main.py
|
||||||
|
[Clip_diffusionDB_ghost]: https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/
|
||||||
|
|
||||||
|
|
||||||
|
[Clip_youtube_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search
|
||||||
|
[Clip_youtube_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search/main.ipynb
|
||||||
|
[Clip_youtube_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search/main.py
|
||||||
|
[Clip_youtube_ghost]: https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/
|
||||||
85
docs/src/examples/python_examples/rag.md
Normal file
85
docs/src/examples/python_examples/rag.md
Normal file
@@ -0,0 +1,85 @@
|
|||||||
|
|
||||||
|
**🔍💡 RAG: Revolutionize Information Retrieval with LanceDB 🔓**
|
||||||
|
====================================================================
|
||||||
|
|
||||||
|
Unlock the full potential of Retrieval-Augmented Generation (RAG) with LanceDB, the ultimate solution for efficient vector-based information retrieval 📊. Input text queries and retrieve relevant documents with lightning-fast speed ⚡️ and accuracy ✅. Generate comprehensive answers by combining retrieved information, uncovering new insights 🔍 and connections.
|
||||||
|
|
||||||
|
### Experience the Future of Search 🔄
|
||||||
|
|
||||||
|
Experience the future of search with RAG, transforming information retrieval and answer generation. Apply RAG to various industries, streamlining processes 📈, saving time ⏰, and resources 💰. Stay ahead of the curve with innovative technology 🔝, powered by LanceDB. Discover the power of RAG with LanceDB and transform your industry with innovative solutions 💡.
|
||||||
|
|
||||||
|
|
||||||
|
| **RAG** | **Description** | **Links** |
|
||||||
|
|----------------------------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------|----------------------------|
|
||||||
|
| **RAG with Matryoshka Embeddings and LlamaIndex** 🪆🔗 | Utilize **Matryoshka embeddings** and **LlamaIndex** to improve the efficiency and accuracy of your RAG models. 📈✨ | [][matryoshka_github] <br>[][matryoshka_colab] |
|
||||||
|
| **Improve RAG with Re-ranking** 📈🔄 | Enhance your RAG applications by implementing **re-ranking strategies** for more relevant document retrieval. 📚🔍 | [][rag_reranking_github] <br>[][rag_reranking_colab] <br>[][rag_reranking_ghost] |
|
||||||
|
| **Instruct-Multitask** 🧠🎯 | Integrate the **Instruct Embedding Model** with LanceDB to streamline your embedding API, reducing redundant code and overhead. 🌐📊 | [][instruct_multitask_github] <br>[][instruct_multitask_colab] <br>[][instruct_multitask_python] <br>[][instruct_multitask_ghost] |
|
||||||
|
| **Improve RAG with HyDE** 🌌🔍 | Use **Hypothetical Document Embeddings** for efficient, accurate, and unsupervised dense retrieval. 📄🔍 | [][hyde_github] <br>[][hyde_colab]<br>[][hyde_ghost] |
|
||||||
|
| **Improve RAG with LOTR** 🧙♂️📜 | Enhance RAG with **Lord of the Retriever (LOTR)** to address 'Lost in the Middle' challenges, especially in medical data. 🌟📜 | [][lotr_github] <br>[][lotr_colab] <br>[][lotr_ghost] |
|
||||||
|
| **Advanced RAG: Parent Document Retriever** 📑🔗 | Use **Parent Document & Bigger Chunk Retriever** to maintain context and relevance when generating related content. 🎵📄 | [][parent_doc_retriever_github] <br>[][parent_doc_retriever_colab] <br>[][parent_doc_retriever_ghost] |
|
||||||
|
| **Corrective RAG with Langgraph** 🔧📊 | Enhance RAG reliability with **Corrective RAG (CRAG)** by self-reflecting and fact-checking for accurate and trustworthy results. ✅🔍 |[][corrective_rag_github] <br>[][corrective_rag_colab] <br>[][corrective_rag_ghost] |
|
||||||
|
| **Contextual Compression with RAG** 🗜️🧠 | Apply **contextual compression techniques** to condense large documents while retaining essential information. 📄🗜️ | [][compression_rag_github] <br>[][compression_rag_colab] <br>[][compression_rag_ghost] |
|
||||||
|
| **Improve RAG with FLARE** 🔥| Enable users to ask questions directly to academic papers, focusing on ArXiv papers, with Forward-Looking Active REtrieval augmented generation.🚀🌟 | [][flare_github] <br>[][flare_colab] <br>[][flare_ghost] |
|
||||||
|
| **Query Expansion and Reranker** 🔍🔄 | Enhance RAG with query expansion using Large Language Models and advanced **reranking methods** like Cross Encoders, ColBERT v2, and FlashRank for improved document retrieval precision and recall 🔍📈 | [][query_github] <br>[][query_colab] |
|
||||||
|
| **RAG Fusion** ⚡🌐 | Revolutionize search with RAG Fusion, utilizing the **RRF algorithm** to rerank documents based on user queries, and leveraging LanceDB and OPENAI Embeddings for efficient information retrieval ⚡🌐 | [][fusion_github] <br>[][fusion_colab] |
|
||||||
|
| **Agentic RAG** 🤖📚 | Unlock autonomous information retrieval with **Agentic RAG**, a framework of **intelligent agents** that collaborate to synthesize, summarize, and compare data across sources, enabling proactive and informed decision-making 🤖📚 | [][agentic_github] <br>[][agentic_colab] |
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
[matryoshka_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/RAG-with_MatryoshkaEmbed-Llamaindex
|
||||||
|
[matryoshka_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/RAG-with_MatryoshkaEmbed-Llamaindex/RAG_with_MatryoshkaEmbedding_and_Llamaindex.ipynb
|
||||||
|
|
||||||
|
[rag_reranking_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/RAG_Reranking
|
||||||
|
[rag_reranking_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/RAG_Reranking/main.ipynb
|
||||||
|
[rag_reranking_ghost]: https://blog.lancedb.com/simplest-method-to-improve-rag-pipeline-re-ranking-cf6eaec6d544
|
||||||
|
|
||||||
|
|
||||||
|
[instruct_multitask_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask
|
||||||
|
[instruct_multitask_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask/main.ipynb
|
||||||
|
[instruct_multitask_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask/main.py
|
||||||
|
[instruct_multitask_ghost]: https://blog.lancedb.com/multitask-embedding-with-lancedb-be18ec397543
|
||||||
|
|
||||||
|
[hyde_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE
|
||||||
|
[hyde_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE/main.ipynb
|
||||||
|
[hyde_ghost]: https://blog.lancedb.com/advanced-rag-precise-zero-shot-dense-retrieval-with-hyde-0946c54dfdcb
|
||||||
|
|
||||||
|
[lotr_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Advance_RAG_LOTR
|
||||||
|
[lotr_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance_RAG_LOTR/main.ipynb
|
||||||
|
[lotr_ghost]: https://blog.lancedb.com/better-rag-with-lotr-lord-of-retriever-23c8336b9a35
|
||||||
|
|
||||||
|
[parent_doc_retriever_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/parent_document_retriever
|
||||||
|
[parent_doc_retriever_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/parent_document_retriever/main.ipynb
|
||||||
|
[parent_doc_retriever_ghost]: https://blog.lancedb.com/modified-rag-parent-document-bigger-chunk-retriever-62b3d1e79bc6
|
||||||
|
|
||||||
|
[corrective_rag_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph
|
||||||
|
[corrective_rag_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph/CRAG_with_Langgraph.ipynb
|
||||||
|
[corrective_rag_ghost]: https://blog.lancedb.com/implementing-corrective-rag-in-the-easiest-way-2/
|
||||||
|
|
||||||
|
[compression_rag_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Contextual-Compression-with-RAG
|
||||||
|
[compression_rag_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Contextual-Compression-with-RAG/main.ipynb
|
||||||
|
[compression_rag_ghost]: https://blog.lancedb.com/enhance-rag-integrate-contextual-compression-and-filtering-for-precision-a29d4a810301/
|
||||||
|
|
||||||
|
[flare_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR
|
||||||
|
[flare_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR/main.ipynb
|
||||||
|
[flare_ghost]: https://blog.lancedb.com/better-rag-with-active-retrieval-augmented-generation-flare-3b66646e2a9f/
|
||||||
|
|
||||||
|
[query_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/QueryExpansion&Reranker
|
||||||
|
[query_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/QueryExpansion&Reranker/main.ipynb
|
||||||
|
|
||||||
|
|
||||||
|
[fusion_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/RAG_Fusion
|
||||||
|
[fusion_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/RAG_Fusion/main.ipynb
|
||||||
|
|
||||||
|
[agentic_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG
|
||||||
|
[agentic_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG/main.ipynb
|
||||||
|
|
||||||
|
|
||||||
167
docs/src/fts.md
167
docs/src/fts.md
@@ -1,9 +1,14 @@
|
|||||||
# Full-text search
|
# Full-text search
|
||||||
|
|
||||||
LanceDB provides support for full-text search via [Tantivy](https://github.com/quickwit-oss/tantivy) (currently Python only), allowing you to incorporate keyword-based search (based on BM25) in your retrieval solutions. Our goal is to push the FTS integration down to the Rust level in the future, so that it's available for Rust and JavaScript users as well. Follow along at [this Github issue](https://github.com/lancedb/lance/issues/1195)
|
LanceDB provides support for full-text search via Lance (before via [Tantivy](https://github.com/quickwit-oss/tantivy) (Python only)), allowing you to incorporate keyword-based search (based on BM25) in your retrieval solutions.
|
||||||
|
|
||||||
|
Currently, the Lance full text search is missing some features that are in the Tantivy full text search. This includes phrase queries, re-ranking, and customizing the tokenizer. Thus, in Python, Tantivy is still the default way to do full text search and many of the instructions below apply just to Tantivy-based indices.
|
||||||
|
|
||||||
|
|
||||||
## Installation
|
## Installation (Only for Tantivy-based FTS)
|
||||||
|
|
||||||
|
!!! note
|
||||||
|
No need to install the tantivy dependency if using native FTS
|
||||||
|
|
||||||
To use full-text search, install the dependency [`tantivy-py`](https://github.com/quickwit-oss/tantivy-py):
|
To use full-text search, install the dependency [`tantivy-py`](https://github.com/quickwit-oss/tantivy-py):
|
||||||
|
|
||||||
@@ -14,42 +19,83 @@ pip install tantivy==0.20.1
|
|||||||
|
|
||||||
## Example
|
## Example
|
||||||
|
|
||||||
Consider that we have a LanceDB table named `my_table`, whose string column `text` we want to index and query via keyword search.
|
Consider that we have a LanceDB table named `my_table`, whose string column `text` we want to index and query via keyword search, the FTS index must be created before you can search via keywords.
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
import lancedb
|
|
||||||
|
|
||||||
uri = "data/sample-lancedb"
|
```python
|
||||||
db = lancedb.connect(uri)
|
import lancedb
|
||||||
|
|
||||||
table = db.create_table(
|
uri = "data/sample-lancedb"
|
||||||
|
db = lancedb.connect(uri)
|
||||||
|
|
||||||
|
table = db.create_table(
|
||||||
"my_table",
|
"my_table",
|
||||||
data=[
|
data=[
|
||||||
{"vector": [3.1, 4.1], "text": "Frodo was a happy puppy"},
|
{"vector": [3.1, 4.1], "text": "Frodo was a happy puppy"},
|
||||||
{"vector": [5.9, 26.5], "text": "There are several kittens playing"},
|
{"vector": [5.9, 26.5], "text": "There are several kittens playing"},
|
||||||
],
|
],
|
||||||
)
|
)
|
||||||
```
|
|
||||||
|
|
||||||
## Create FTS index on single column
|
# passing `use_tantivy=False` to use lance FTS index
|
||||||
|
# `use_tantivy=True` by default
|
||||||
|
table.create_fts_index("text")
|
||||||
|
table.search("puppy").limit(10).select(["text"]).to_list()
|
||||||
|
# [{'text': 'Frodo was a happy puppy', '_score': 0.6931471824645996}]
|
||||||
|
# ...
|
||||||
|
```
|
||||||
|
|
||||||
The FTS index must be created before you can search via keywords.
|
=== "TypeScript"
|
||||||
|
|
||||||
```python
|
```typescript
|
||||||
table.create_fts_index("text")
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
```
|
const uri = "data/sample-lancedb"
|
||||||
|
const db = await lancedb.connect(uri);
|
||||||
|
|
||||||
To search an FTS index via keywords, LanceDB's `table.search` accepts a string as input:
|
const data = [
|
||||||
|
{ vector: [3.1, 4.1], text: "Frodo was a happy puppy" },
|
||||||
|
{ vector: [5.9, 26.5], text: "There are several kittens playing" },
|
||||||
|
];
|
||||||
|
const tbl = await db.createTable("my_table", data, { mode: "overwrite" });
|
||||||
|
await tbl.createIndex("text", {
|
||||||
|
config: lancedb.Index.fts(),
|
||||||
|
});
|
||||||
|
|
||||||
```python
|
await tbl
|
||||||
table.search("puppy").limit(10).select(["text"]).to_list()
|
.search("puppy")
|
||||||
```
|
.select(["text"])
|
||||||
|
.limit(10)
|
||||||
|
.toArray();
|
||||||
|
```
|
||||||
|
|
||||||
This returns the result as a list of dictionaries as follows.
|
=== "Rust"
|
||||||
|
|
||||||
```python
|
```rust
|
||||||
[{'text': 'Frodo was a happy puppy', 'score': 0.6931471824645996}]
|
let uri = "data/sample-lancedb";
|
||||||
```
|
let db = connect(uri).execute().await?;
|
||||||
|
let initial_data: Box<dyn RecordBatchReader + Send> = create_some_records()?;
|
||||||
|
let tbl = db
|
||||||
|
.create_table("my_table", initial_data)
|
||||||
|
.execute()
|
||||||
|
.await?;
|
||||||
|
tbl
|
||||||
|
.create_index(&["text"], Index::FTS(FtsIndexBuilder::default()))
|
||||||
|
.execute()
|
||||||
|
.await?;
|
||||||
|
|
||||||
|
tbl
|
||||||
|
.query()
|
||||||
|
.full_text_search(FullTextSearchQuery::new("puppy".to_owned()))
|
||||||
|
.select(lancedb::query::Select::Columns(vec!["text".to_owned()]))
|
||||||
|
.limit(10)
|
||||||
|
.execute()
|
||||||
|
.await?;
|
||||||
|
```
|
||||||
|
|
||||||
|
It would search on all indexed columns by default, so it's useful when there are multiple indexed columns.
|
||||||
|
For now, this is supported in tantivy way only.
|
||||||
|
|
||||||
|
Passing `fts_columns="text"` if you want to specify the columns to search, but it's not available for Tantivy-based full text search.
|
||||||
|
|
||||||
!!! note
|
!!! note
|
||||||
LanceDB automatically searches on the existing FTS index if the input to the search is of type `str`. If you provide a vector as input, LanceDB will search the ANN index instead.
|
LanceDB automatically searches on the existing FTS index if the input to the search is of type `str`. If you provide a vector as input, LanceDB will search the ANN index instead.
|
||||||
@@ -57,20 +103,33 @@ This returns the result as a list of dictionaries as follows.
|
|||||||
## Tokenization
|
## Tokenization
|
||||||
By default the text is tokenized by splitting on punctuation and whitespaces and then removing tokens that are longer than 40 chars. For more language specific tokenization then provide the argument tokenizer_name with the 2 letter language code followed by "_stem". So for english it would be "en_stem".
|
By default the text is tokenized by splitting on punctuation and whitespaces and then removing tokens that are longer than 40 chars. For more language specific tokenization then provide the argument tokenizer_name with the 2 letter language code followed by "_stem". So for english it would be "en_stem".
|
||||||
|
|
||||||
```python
|
For now, only the Tantivy-based FTS index supports to specify the tokenizer, so it's only available in Python with `use_tantivy=True`.
|
||||||
table.create_fts_index("text", tokenizer_name="en_stem")
|
|
||||||
```
|
|
||||||
|
|
||||||
The following [languages](https://docs.rs/tantivy/latest/tantivy/tokenizer/enum.Language.html) are currently supported.
|
=== "use_tantivy=True"
|
||||||
|
|
||||||
|
```python
|
||||||
|
table.create_fts_index("text", use_tantivy=True, tokenizer_name="en_stem")
|
||||||
|
```
|
||||||
|
|
||||||
|
=== "use_tantivy=False"
|
||||||
|
|
||||||
|
[**Not supported yet**](https://github.com/lancedb/lance/issues/1195)
|
||||||
|
|
||||||
|
the following [languages](https://docs.rs/tantivy/latest/tantivy/tokenizer/enum.Language.html) are currently supported.
|
||||||
|
|
||||||
## Index multiple columns
|
## Index multiple columns
|
||||||
|
|
||||||
If you have multiple string columns to index, there's no need to combine them manually -- simply pass them all as a list to `create_fts_index`:
|
If you have multiple string columns to index, there's no need to combine them manually -- simply pass them all as a list to `create_fts_index`:
|
||||||
|
|
||||||
```python
|
=== "use_tantivy=True"
|
||||||
table.create_fts_index(["text1", "text2"])
|
|
||||||
```
|
```python
|
||||||
|
table.create_fts_index(["text1", "text2"])
|
||||||
|
```
|
||||||
|
|
||||||
|
=== "use_tantivy=False"
|
||||||
|
|
||||||
|
[**Not supported yet**](https://github.com/lancedb/lance/issues/1195)
|
||||||
|
|
||||||
Note that the search API call does not change - you can search over all indexed columns at once.
|
Note that the search API call does not change - you can search over all indexed columns at once.
|
||||||
|
|
||||||
@@ -80,19 +139,48 @@ Currently the LanceDB full text search feature supports *post-filtering*, meanin
|
|||||||
applied on top of the full text search results. This can be invoked via the familiar
|
applied on top of the full text search results. This can be invoked via the familiar
|
||||||
`where` syntax:
|
`where` syntax:
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
table.search("puppy").limit(10).where("meta='foo'").to_list()
|
|
||||||
```
|
```python
|
||||||
|
table.search("puppy").limit(10).where("meta='foo'").to_list()
|
||||||
|
```
|
||||||
|
|
||||||
|
=== "TypeScript"
|
||||||
|
|
||||||
|
```typescript
|
||||||
|
await tbl
|
||||||
|
.search("apple")
|
||||||
|
.select(["id", "doc"])
|
||||||
|
.limit(10)
|
||||||
|
.where("meta='foo'")
|
||||||
|
.toArray();
|
||||||
|
```
|
||||||
|
|
||||||
|
=== "Rust"
|
||||||
|
|
||||||
|
```rust
|
||||||
|
table
|
||||||
|
.query()
|
||||||
|
.full_text_search(FullTextSearchQuery::new(words[0].to_owned()))
|
||||||
|
.select(lancedb::query::Select::Columns(vec!["doc".to_owned()]))
|
||||||
|
.limit(10)
|
||||||
|
.only_if("meta='foo'")
|
||||||
|
.execute()
|
||||||
|
.await?;
|
||||||
|
```
|
||||||
|
|
||||||
## Sorting
|
## Sorting
|
||||||
|
|
||||||
|
!!! warning "Warn"
|
||||||
|
Sorting is available for only Tantivy-based FTS
|
||||||
|
|
||||||
You can pre-sort the documents by specifying `ordering_field_names` when
|
You can pre-sort the documents by specifying `ordering_field_names` when
|
||||||
creating the full-text search index. Once pre-sorted, you can then specify
|
creating the full-text search index. Once pre-sorted, you can then specify
|
||||||
`ordering_field_name` while searching to return results sorted by the given
|
`ordering_field_name` while searching to return results sorted by the given
|
||||||
field. For example,
|
field. For example,
|
||||||
|
|
||||||
```
|
```python
|
||||||
table.create_fts_index(["text_field"], ordering_field_names=["sort_by_field"])
|
table.create_fts_index(["text_field"], use_tantivy=True, ordering_field_names=["sort_by_field"])
|
||||||
|
|
||||||
(table.search("terms", ordering_field_name="sort_by_field")
|
(table.search("terms", ordering_field_name="sort_by_field")
|
||||||
.limit(20)
|
.limit(20)
|
||||||
@@ -116,6 +204,9 @@ table.create_fts_index(["text_field"], ordering_field_names=["sort_by_field"])
|
|||||||
|
|
||||||
## Phrase queries vs. terms queries
|
## Phrase queries vs. terms queries
|
||||||
|
|
||||||
|
!!! warning "Warn"
|
||||||
|
Phrase queries are available for only Tantivy-based FTS
|
||||||
|
|
||||||
For full-text search you can specify either a **phrase** query like `"the old man and the sea"`,
|
For full-text search you can specify either a **phrase** query like `"the old man and the sea"`,
|
||||||
or a **terms** search query like `"(Old AND Man) AND Sea"`. For more details on the terms
|
or a **terms** search query like `"(Old AND Man) AND Sea"`. For more details on the terms
|
||||||
query syntax, see Tantivy's [query parser rules](https://docs.rs/tantivy/latest/tantivy/query/struct.QueryParser.html).
|
query syntax, see Tantivy's [query parser rules](https://docs.rs/tantivy/latest/tantivy/query/struct.QueryParser.html).
|
||||||
@@ -142,7 +233,7 @@ enforce it in one of two ways:
|
|||||||
|
|
||||||
1. Place the double-quoted query inside single quotes. For example, `table.search('"they could have been dogs OR cats"')` is treated as
|
1. Place the double-quoted query inside single quotes. For example, `table.search('"they could have been dogs OR cats"')` is treated as
|
||||||
a phrase query.
|
a phrase query.
|
||||||
2. Explicitly declare the `phrase_query()` method. This is useful when you have a phrase query that
|
1. Explicitly declare the `phrase_query()` method. This is useful when you have a phrase query that
|
||||||
itself contains double quotes. For example, `table.search('the cats OR dogs were not really "pets" at all').phrase_query()`
|
itself contains double quotes. For example, `table.search('the cats OR dogs were not really "pets" at all').phrase_query()`
|
||||||
is treated as a phrase query.
|
is treated as a phrase query.
|
||||||
|
|
||||||
@@ -150,7 +241,7 @@ In general, a query that's declared as a phrase query will be wrapped in double
|
|||||||
double quotes replaced by single quotes.
|
double quotes replaced by single quotes.
|
||||||
|
|
||||||
|
|
||||||
## Configurations
|
## Configurations (Only for Tantivy-based FTS)
|
||||||
|
|
||||||
By default, LanceDB configures a 1GB heap size limit for creating the index. You can
|
By default, LanceDB configures a 1GB heap size limit for creating the index. You can
|
||||||
reduce this if running on a smaller node, or increase this for faster performance while
|
reduce this if running on a smaller node, or increase this for faster performance while
|
||||||
@@ -164,6 +255,8 @@ table.create_fts_index(["text1", "text2"], writer_heap_size=heap, replace=True)
|
|||||||
|
|
||||||
## Current limitations
|
## Current limitations
|
||||||
|
|
||||||
|
For that Tantivy-based FTS:
|
||||||
|
|
||||||
1. Currently we do not yet support incremental writes.
|
1. Currently we do not yet support incremental writes.
|
||||||
If you add data after FTS index creation, it won't be reflected
|
If you add data after FTS index creation, it won't be reflected
|
||||||
in search results until you do a full reindex.
|
in search results until you do a full reindex.
|
||||||
|
|||||||
File diff suppressed because one or more lines are too long
@@ -113,6 +113,10 @@ lists the indices that LanceDb supports.
|
|||||||
|
|
||||||
::: lancedb.index.BTree
|
::: lancedb.index.BTree
|
||||||
|
|
||||||
|
::: lancedb.index.Bitmap
|
||||||
|
|
||||||
|
::: lancedb.index.LabelList
|
||||||
|
|
||||||
::: lancedb.index.IvfPq
|
::: lancedb.index.IvfPq
|
||||||
|
|
||||||
## Querying (Asynchronous)
|
## Querying (Asynchronous)
|
||||||
|
|||||||
53
docs/src/reranking/rrf.md
Normal file
53
docs/src/reranking/rrf.md
Normal file
@@ -0,0 +1,53 @@
|
|||||||
|
# Reciprocal Rank Fusion Reranker
|
||||||
|
|
||||||
|
Reciprocal Rank Fusion (RRF) is an algorithm that evaluates the search scores by leveraging the positions/rank of the documents. The implementation follows this [paper](https://plg.uwaterloo.ca/~gvcormac/cormacksigir09-rrf.pdf).
|
||||||
|
|
||||||
|
|
||||||
|
!!! note
|
||||||
|
Supported Query Types: Hybrid
|
||||||
|
|
||||||
|
|
||||||
|
```python
|
||||||
|
import numpy
|
||||||
|
import lancedb
|
||||||
|
from lancedb.embeddings import get_registry
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.rerankers import RRFReranker
|
||||||
|
|
||||||
|
embedder = get_registry().get("sentence-transformers").create()
|
||||||
|
db = lancedb.connect("~/.lancedb")
|
||||||
|
|
||||||
|
class Schema(LanceModel):
|
||||||
|
text: str = embedder.SourceField()
|
||||||
|
vector: Vector(embedder.ndims()) = embedder.VectorField()
|
||||||
|
|
||||||
|
data = [
|
||||||
|
{"text": "hello world"},
|
||||||
|
{"text": "goodbye world"}
|
||||||
|
]
|
||||||
|
tbl = db.create_table("test", schema=Schema, mode="overwrite")
|
||||||
|
tbl.add(data)
|
||||||
|
reranker = RRFReranker()
|
||||||
|
|
||||||
|
# Run hybrid search with a reranker
|
||||||
|
tbl.create_fts_index("text", replace=True)
|
||||||
|
result = tbl.search("hello", query_type="hybrid").rerank(reranker=reranker).to_list()
|
||||||
|
|
||||||
|
```
|
||||||
|
|
||||||
|
Accepted Arguments
|
||||||
|
----------------
|
||||||
|
| Argument | Type | Default | Description |
|
||||||
|
| --- | --- | --- | --- |
|
||||||
|
| `K` | `int` | `60` | A constant used in the RRF formula (default is 60). Experiments indicate that k = 60 was near-optimal, but that the choice is not critical |
|
||||||
|
| `return_score` | str | `"relevance"` | Options are "relevance" or "all". The type of score to return. If "relevance", will return only the `_relevance_score`. If "all", will return all scores from the vector and FTS search along with the relevance score. |
|
||||||
|
|
||||||
|
|
||||||
|
## Supported Scores for each query type
|
||||||
|
You can specify the type of scores you want the reranker to return. The following are the supported scores for each query type:
|
||||||
|
|
||||||
|
### Hybrid Search
|
||||||
|
|`return_score`| Status | Description |
|
||||||
|
| --- | --- | --- |
|
||||||
|
| `relevance` | ✅ Supported | Returned rows only have the `_relevance_score` column |
|
||||||
|
| `all` | ✅ Supported | Returned rows have vector(`_distance`) and FTS(`score`) along with Hybrid Search score(`_relevance_score`) |
|
||||||
@@ -5,4 +5,5 @@ pylance
|
|||||||
duckdb
|
duckdb
|
||||||
--extra-index-url https://download.pytorch.org/whl/cpu
|
--extra-index-url https://download.pytorch.org/whl/cpu
|
||||||
torch
|
torch
|
||||||
polars
|
polars>=0.19, <=1.3.0
|
||||||
|
|
||||||
|
|||||||
4
node/package-lock.json
generated
4
node/package-lock.json
generated
@@ -1,12 +1,12 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"lockfileVersion": 3,
|
"lockfileVersion": 3,
|
||||||
"requires": true,
|
"requires": true,
|
||||||
"packages": {
|
"packages": {
|
||||||
"": {
|
"": {
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"cpu": [
|
"cpu": [
|
||||||
"x64",
|
"x64",
|
||||||
"arm64"
|
"arm64"
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"description": " Serverless, low-latency vector database for AI applications",
|
"description": " Serverless, low-latency vector database for AI applications",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"types": "dist/index.d.ts",
|
"types": "dist/index.d.ts",
|
||||||
|
|||||||
@@ -13,3 +13,13 @@ __test__
|
|||||||
renovate.json
|
renovate.json
|
||||||
.idea
|
.idea
|
||||||
src
|
src
|
||||||
|
lancedb
|
||||||
|
examples
|
||||||
|
nodejs-artifacts
|
||||||
|
Cargo.toml
|
||||||
|
biome.json
|
||||||
|
build.rs
|
||||||
|
jest.config.js
|
||||||
|
native.d.ts
|
||||||
|
tsconfig.json
|
||||||
|
typedoc.json
|
||||||
@@ -20,7 +20,6 @@ napi = { version = "2.16.8", default-features = false, features = [
|
|||||||
"async",
|
"async",
|
||||||
] }
|
] }
|
||||||
napi-derive = "2.16.4"
|
napi-derive = "2.16.4"
|
||||||
|
|
||||||
# Prevent dynamic linking of lzma, which comes from datafusion
|
# Prevent dynamic linking of lzma, which comes from datafusion
|
||||||
lzma-sys = { version = "*", features = ["static"] }
|
lzma-sys = { version = "*", features = ["static"] }
|
||||||
|
|
||||||
|
|||||||
@@ -1,3 +1,4 @@
|
|||||||
|
import * as apiArrow from "apache-arrow";
|
||||||
// Copyright 2024 Lance Developers.
|
// Copyright 2024 Lance Developers.
|
||||||
//
|
//
|
||||||
// Licensed under the Apache License, Version 2.0 (the "License");
|
// Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
@@ -69,7 +70,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -82,7 +83,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
|
|
||||||
const schema = LanceSchema({
|
const schema = LanceSchema({
|
||||||
id: new arrow.Int32(),
|
id: new arrow.Int32(),
|
||||||
text: func.sourceField(new arrow.Utf8()),
|
text: func.sourceField(new arrow.Utf8() as apiArrow.DataType),
|
||||||
vector: func.vectorField(),
|
vector: func.vectorField(),
|
||||||
});
|
});
|
||||||
|
|
||||||
@@ -119,7 +120,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -144,7 +145,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -154,7 +155,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
|
|
||||||
const schema = LanceSchema({
|
const schema = LanceSchema({
|
||||||
id: new arrow.Int32(),
|
id: new arrow.Int32(),
|
||||||
text: func.sourceField(new arrow.Utf8()),
|
text: func.sourceField(new arrow.Utf8() as apiArrow.DataType),
|
||||||
vector: func.vectorField(),
|
vector: func.vectorField(),
|
||||||
});
|
});
|
||||||
const expectedMetadata = new Map<string, string>([
|
const expectedMetadata = new Map<string, string>([
|
||||||
|
|||||||
@@ -31,7 +31,9 @@ import {
|
|||||||
Float64,
|
Float64,
|
||||||
Int32,
|
Int32,
|
||||||
Int64,
|
Int64,
|
||||||
|
List,
|
||||||
Schema,
|
Schema,
|
||||||
|
Utf8,
|
||||||
makeArrowTable,
|
makeArrowTable,
|
||||||
} from "../lancedb/arrow";
|
} from "../lancedb/arrow";
|
||||||
import {
|
import {
|
||||||
@@ -331,6 +333,7 @@ describe("When creating an index", () => {
|
|||||||
const schema = new Schema([
|
const schema = new Schema([
|
||||||
new Field("id", new Int32(), true),
|
new Field("id", new Int32(), true),
|
||||||
new Field("vec", new FixedSizeList(32, new Field("item", new Float32()))),
|
new Field("vec", new FixedSizeList(32, new Field("item", new Float32()))),
|
||||||
|
new Field("tags", new List(new Field("item", new Utf8(), true))),
|
||||||
]);
|
]);
|
||||||
let tbl: Table;
|
let tbl: Table;
|
||||||
let queryVec: number[];
|
let queryVec: number[];
|
||||||
@@ -346,6 +349,7 @@ describe("When creating an index", () => {
|
|||||||
vec: Array(32)
|
vec: Array(32)
|
||||||
.fill(1)
|
.fill(1)
|
||||||
.map(() => Math.random()),
|
.map(() => Math.random()),
|
||||||
|
tags: ["tag1", "tag2", "tag3"],
|
||||||
})),
|
})),
|
||||||
{
|
{
|
||||||
schema,
|
schema,
|
||||||
@@ -428,6 +432,22 @@ describe("When creating an index", () => {
|
|||||||
}
|
}
|
||||||
});
|
});
|
||||||
|
|
||||||
|
test("create a bitmap index", async () => {
|
||||||
|
await tbl.createIndex("id", {
|
||||||
|
config: Index.bitmap(),
|
||||||
|
});
|
||||||
|
const indexDir = path.join(tmpDir.name, "test.lance", "_indices");
|
||||||
|
expect(fs.readdirSync(indexDir)).toHaveLength(1);
|
||||||
|
});
|
||||||
|
|
||||||
|
test("create a label list index", async () => {
|
||||||
|
await tbl.createIndex("tags", {
|
||||||
|
config: Index.labelList(),
|
||||||
|
});
|
||||||
|
const indexDir = path.join(tmpDir.name, "test.lance", "_indices");
|
||||||
|
expect(fs.readdirSync(indexDir)).toHaveLength(1);
|
||||||
|
});
|
||||||
|
|
||||||
test("should be able to get index stats", async () => {
|
test("should be able to get index stats", async () => {
|
||||||
await tbl.createIndex("id");
|
await tbl.createIndex("id");
|
||||||
|
|
||||||
@@ -785,11 +805,26 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
];
|
];
|
||||||
const table = await db.createTable("test", data);
|
const table = await db.createTable("test", data);
|
||||||
|
|
||||||
expect(table.search("hello").toArray()).rejects.toThrow(
|
expect(table.search("hello", "vector").toArray()).rejects.toThrow(
|
||||||
"No embedding functions are defined in the table",
|
"No embedding functions are defined in the table",
|
||||||
);
|
);
|
||||||
});
|
});
|
||||||
|
|
||||||
|
test("full text search if no embedding function provided", async () => {
|
||||||
|
const db = await connect(tmpDir.name);
|
||||||
|
const data = [
|
||||||
|
{ text: "hello world", vector: [0.1, 0.2, 0.3] },
|
||||||
|
{ text: "goodbye world", vector: [0.4, 0.5, 0.6] },
|
||||||
|
];
|
||||||
|
const table = await db.createTable("test", data);
|
||||||
|
await table.createIndex("text", {
|
||||||
|
config: Index.fts(),
|
||||||
|
});
|
||||||
|
|
||||||
|
const results = await table.search("hello").toArray();
|
||||||
|
expect(results[0].text).toBe(data[0].text);
|
||||||
|
});
|
||||||
|
|
||||||
test.each([
|
test.each([
|
||||||
[0.4, 0.5, 0.599], // number[]
|
[0.4, 0.5, 0.599], // number[]
|
||||||
Float32Array.of(0.4, 0.5, 0.599), // Float32Array
|
Float32Array.of(0.4, 0.5, 0.599), // Float32Array
|
||||||
|
|||||||
64
nodejs/examples/custom_embedding_function.ts
Normal file
64
nodejs/examples/custom_embedding_function.ts
Normal file
@@ -0,0 +1,64 @@
|
|||||||
|
// --8<-- [start:imports]
|
||||||
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
|
import {
|
||||||
|
LanceSchema,
|
||||||
|
TextEmbeddingFunction,
|
||||||
|
getRegistry,
|
||||||
|
register,
|
||||||
|
} from "@lancedb/lancedb/embedding";
|
||||||
|
import { pipeline } from "@xenova/transformers";
|
||||||
|
// --8<-- [end:imports]
|
||||||
|
|
||||||
|
// --8<-- [start:embedding_impl]
|
||||||
|
@register("sentence-transformers")
|
||||||
|
class SentenceTransformersEmbeddings extends TextEmbeddingFunction {
|
||||||
|
name = "Xenova/all-miniLM-L6-v2";
|
||||||
|
#ndims!: number;
|
||||||
|
extractor: any;
|
||||||
|
|
||||||
|
async init() {
|
||||||
|
this.extractor = await pipeline("feature-extraction", this.name);
|
||||||
|
this.#ndims = await this.generateEmbeddings(["hello"]).then(
|
||||||
|
(e) => e[0].length,
|
||||||
|
);
|
||||||
|
}
|
||||||
|
|
||||||
|
ndims() {
|
||||||
|
return this.#ndims;
|
||||||
|
}
|
||||||
|
|
||||||
|
toJSON() {
|
||||||
|
return {
|
||||||
|
name: this.name,
|
||||||
|
};
|
||||||
|
}
|
||||||
|
async generateEmbeddings(texts: string[]) {
|
||||||
|
const output = await this.extractor(texts, {
|
||||||
|
pooling: "mean",
|
||||||
|
normalize: true,
|
||||||
|
});
|
||||||
|
return output.tolist();
|
||||||
|
}
|
||||||
|
}
|
||||||
|
// -8<-- [end:embedding_impl]
|
||||||
|
|
||||||
|
// --8<-- [start:call_custom_function]
|
||||||
|
const registry = getRegistry();
|
||||||
|
|
||||||
|
const sentenceTransformer = await registry
|
||||||
|
.get<SentenceTransformersEmbeddings>("sentence-transformers")!
|
||||||
|
.create();
|
||||||
|
|
||||||
|
const schema = LanceSchema({
|
||||||
|
vector: sentenceTransformer.vectorField(),
|
||||||
|
text: sentenceTransformer.sourceField(),
|
||||||
|
});
|
||||||
|
|
||||||
|
const db = await lancedb.connect("/tmp/db");
|
||||||
|
const table = await db.createEmptyTable("table", schema, { mode: "overwrite" });
|
||||||
|
|
||||||
|
await table.add([{ text: "hello" }, { text: "world" }]);
|
||||||
|
|
||||||
|
const results = await table.search("greeting").limit(1).toArray();
|
||||||
|
console.log(results[0].text);
|
||||||
|
// -8<-- [end:call_custom_function]
|
||||||
52
nodejs/examples/full_text_search.ts
Normal file
52
nodejs/examples/full_text_search.ts
Normal file
@@ -0,0 +1,52 @@
|
|||||||
|
// Copyright 2024 Lance Developers.
|
||||||
|
//
|
||||||
|
// Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
|
// you may not use this file except in compliance with the License.
|
||||||
|
// You may obtain a copy of the License at
|
||||||
|
//
|
||||||
|
// http://www.apache.org/licenses/LICENSE-2.0
|
||||||
|
//
|
||||||
|
// Unless required by applicable law or agreed to in writing, software
|
||||||
|
// distributed under the License is distributed on an "AS IS" BASIS,
|
||||||
|
// WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
||||||
|
// See the License for the specific language governing permissions and
|
||||||
|
// limitations under the License.
|
||||||
|
|
||||||
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
|
|
||||||
|
const db = await lancedb.connect("data/sample-lancedb");
|
||||||
|
|
||||||
|
const words = [
|
||||||
|
"apple",
|
||||||
|
"banana",
|
||||||
|
"cherry",
|
||||||
|
"date",
|
||||||
|
"elderberry",
|
||||||
|
"fig",
|
||||||
|
"grape",
|
||||||
|
];
|
||||||
|
|
||||||
|
const data = Array.from({ length: 10_000 }, (_, i) => ({
|
||||||
|
vector: Array(1536).fill(i),
|
||||||
|
id: i,
|
||||||
|
item: `item ${i}`,
|
||||||
|
strId: `${i}`,
|
||||||
|
doc: words[i % words.length],
|
||||||
|
}));
|
||||||
|
|
||||||
|
const tbl = await db.createTable("myVectors", data, { mode: "overwrite" });
|
||||||
|
|
||||||
|
await tbl.createIndex("doc", {
|
||||||
|
config: lancedb.Index.fts(),
|
||||||
|
});
|
||||||
|
|
||||||
|
// --8<-- [start:full_text_search]
|
||||||
|
let result = await tbl
|
||||||
|
.search("apple")
|
||||||
|
.select(["id", "doc"])
|
||||||
|
.limit(10)
|
||||||
|
.toArray();
|
||||||
|
console.log(result);
|
||||||
|
// --8<-- [end:full_text_search]
|
||||||
|
|
||||||
|
console.log("SQL search: done");
|
||||||
793
nodejs/examples/package-lock.json
generated
793
nodejs/examples/package-lock.json
generated
@@ -9,7 +9,12 @@
|
|||||||
"version": "1.0.0",
|
"version": "1.0.0",
|
||||||
"license": "Apache-2.0",
|
"license": "Apache-2.0",
|
||||||
"dependencies": {
|
"dependencies": {
|
||||||
"@lancedb/lancedb": "file:../"
|
"@lancedb/lancedb": "file:../",
|
||||||
|
"@xenova/transformers": "^2.17.2",
|
||||||
|
"tsc": "^2.0.4"
|
||||||
|
},
|
||||||
|
"devDependencies": {
|
||||||
|
"typescript": "^5.5.4"
|
||||||
},
|
},
|
||||||
"peerDependencies": {
|
"peerDependencies": {
|
||||||
"typescript": "^5.0.0"
|
"typescript": "^5.0.0"
|
||||||
@@ -17,7 +22,7 @@
|
|||||||
},
|
},
|
||||||
"..": {
|
"..": {
|
||||||
"name": "@lancedb/lancedb",
|
"name": "@lancedb/lancedb",
|
||||||
"version": "0.6.0",
|
"version": "0.8.0",
|
||||||
"cpu": [
|
"cpu": [
|
||||||
"x64",
|
"x64",
|
||||||
"arm64"
|
"arm64"
|
||||||
@@ -29,44 +34,791 @@
|
|||||||
"win32"
|
"win32"
|
||||||
],
|
],
|
||||||
"dependencies": {
|
"dependencies": {
|
||||||
"apache-arrow": "^15.0.0",
|
|
||||||
"axios": "^1.7.2",
|
"axios": "^1.7.2",
|
||||||
"openai": "^4.29.2",
|
|
||||||
"reflect-metadata": "^0.2.2"
|
"reflect-metadata": "^0.2.2"
|
||||||
},
|
},
|
||||||
"devDependencies": {
|
"devDependencies": {
|
||||||
|
"@aws-sdk/client-dynamodb": "^3.33.0",
|
||||||
"@aws-sdk/client-kms": "^3.33.0",
|
"@aws-sdk/client-kms": "^3.33.0",
|
||||||
"@aws-sdk/client-s3": "^3.33.0",
|
"@aws-sdk/client-s3": "^3.33.0",
|
||||||
"@biomejs/biome": "^1.7.3",
|
"@biomejs/biome": "^1.7.3",
|
||||||
"@jest/globals": "^29.7.0",
|
"@jest/globals": "^29.7.0",
|
||||||
"@napi-rs/cli": "^2.18.0",
|
"@napi-rs/cli": "^2.18.3",
|
||||||
"@types/axios": "^0.14.0",
|
"@types/axios": "^0.14.0",
|
||||||
"@types/jest": "^29.1.2",
|
"@types/jest": "^29.1.2",
|
||||||
"@types/tmp": "^0.2.6",
|
"@types/tmp": "^0.2.6",
|
||||||
"apache-arrow-old": "npm:apache-arrow@13.0.0",
|
"apache-arrow-13": "npm:apache-arrow@13.0.0",
|
||||||
|
"apache-arrow-14": "npm:apache-arrow@14.0.0",
|
||||||
|
"apache-arrow-15": "npm:apache-arrow@15.0.0",
|
||||||
|
"apache-arrow-16": "npm:apache-arrow@16.0.0",
|
||||||
|
"apache-arrow-17": "npm:apache-arrow@17.0.0",
|
||||||
"eslint": "^8.57.0",
|
"eslint": "^8.57.0",
|
||||||
"jest": "^29.7.0",
|
"jest": "^29.7.0",
|
||||||
"shx": "^0.3.4",
|
"shx": "^0.3.4",
|
||||||
"tmp": "^0.2.3",
|
"tmp": "^0.2.3",
|
||||||
"ts-jest": "^29.1.2",
|
"ts-jest": "^29.1.2",
|
||||||
"typedoc": "^0.25.7",
|
"typedoc": "^0.26.4",
|
||||||
"typedoc-plugin-markdown": "^3.17.1",
|
"typedoc-plugin-markdown": "^4.2.1",
|
||||||
"typescript": "^5.3.3",
|
"typescript": "^5.5.4",
|
||||||
"typescript-eslint": "^7.1.0"
|
"typescript-eslint": "^7.1.0"
|
||||||
},
|
},
|
||||||
"engines": {
|
"engines": {
|
||||||
"node": ">= 18"
|
"node": ">= 18"
|
||||||
|
},
|
||||||
|
"optionalDependencies": {
|
||||||
|
"@xenova/transformers": ">=2.17 < 3",
|
||||||
|
"openai": "^4.29.2"
|
||||||
|
},
|
||||||
|
"peerDependencies": {
|
||||||
|
"apache-arrow": ">=13.0.0 <=17.0.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/@huggingface/jinja": {
|
||||||
|
"version": "0.2.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@huggingface/jinja/-/jinja-0.2.2.tgz",
|
||||||
|
"integrity": "sha512-/KPde26khDUIPkTGU82jdtTW9UAuvUTumCAbFs/7giR0SxsvZC4hru51PBvpijH6BVkHcROcvZM/lpy5h1jRRA==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=18"
|
||||||
}
|
}
|
||||||
},
|
},
|
||||||
"node_modules/@lancedb/lancedb": {
|
"node_modules/@lancedb/lancedb": {
|
||||||
"resolved": "..",
|
"resolved": "..",
|
||||||
"link": true
|
"link": true
|
||||||
},
|
},
|
||||||
|
"node_modules/@protobufjs/aspromise": {
|
||||||
|
"version": "1.1.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/aspromise/-/aspromise-1.1.2.tgz",
|
||||||
|
"integrity": "sha512-j+gKExEuLmKwvz3OgROXtrJ2UG2x8Ch2YZUxahh+s1F2HZ+wAceUNLkvy6zKCPVRkU++ZWQrdxsUeQXmcg4uoQ=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/base64": {
|
||||||
|
"version": "1.1.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/base64/-/base64-1.1.2.tgz",
|
||||||
|
"integrity": "sha512-AZkcAA5vnN/v4PDqKyMR5lx7hZttPDgClv83E//FMNhR2TMcLUhfRUBHCmSl0oi9zMgDDqRUJkSxO3wm85+XLg=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/codegen": {
|
||||||
|
"version": "2.0.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/codegen/-/codegen-2.0.4.tgz",
|
||||||
|
"integrity": "sha512-YyFaikqM5sH0ziFZCN3xDC7zeGaB/d0IUb9CATugHWbd1FRFwWwt4ld4OYMPWu5a3Xe01mGAULCdqhMlPl29Jg=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/eventemitter": {
|
||||||
|
"version": "1.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/eventemitter/-/eventemitter-1.1.0.tgz",
|
||||||
|
"integrity": "sha512-j9ednRT81vYJ9OfVuXG6ERSTdEL1xVsNgqpkxMsbIabzSo3goCjDIveeGv5d03om39ML71RdmrGNjG5SReBP/Q=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/fetch": {
|
||||||
|
"version": "1.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/fetch/-/fetch-1.1.0.tgz",
|
||||||
|
"integrity": "sha512-lljVXpqXebpsijW71PZaCYeIcE5on1w5DlQy5WH6GLbFryLUrBD4932W/E2BSpfRJWseIL4v/KPgBFxDOIdKpQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"@protobufjs/aspromise": "^1.1.1",
|
||||||
|
"@protobufjs/inquire": "^1.1.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/float": {
|
||||||
|
"version": "1.0.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/float/-/float-1.0.2.tgz",
|
||||||
|
"integrity": "sha512-Ddb+kVXlXst9d+R9PfTIxh1EdNkgoRe5tOX6t01f1lYWOvJnSPDBlG241QLzcyPdoNTsblLUdujGSE4RzrTZGQ=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/inquire": {
|
||||||
|
"version": "1.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/inquire/-/inquire-1.1.0.tgz",
|
||||||
|
"integrity": "sha512-kdSefcPdruJiFMVSbn801t4vFK7KB/5gd2fYvrxhuJYg8ILrmn9SKSX2tZdV6V+ksulWqS7aXjBcRXl3wHoD9Q=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/path": {
|
||||||
|
"version": "1.1.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/path/-/path-1.1.2.tgz",
|
||||||
|
"integrity": "sha512-6JOcJ5Tm08dOHAbdR3GrvP+yUUfkjG5ePsHYczMFLq3ZmMkAD98cDgcT2iA1lJ9NVwFd4tH/iSSoe44YWkltEA=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/pool": {
|
||||||
|
"version": "1.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/pool/-/pool-1.1.0.tgz",
|
||||||
|
"integrity": "sha512-0kELaGSIDBKvcgS4zkjz1PeddatrjYcmMWOlAuAPwAeccUrPHdUqo/J6LiymHHEiJT5NrF1UVwxY14f+fy4WQw=="
|
||||||
|
},
|
||||||
|
"node_modules/@protobufjs/utf8": {
|
||||||
|
"version": "1.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/@protobufjs/utf8/-/utf8-1.1.0.tgz",
|
||||||
|
"integrity": "sha512-Vvn3zZrhQZkkBE8LSuW3em98c0FwgO4nxzv6OdSxPKJIEKY2bGbHn+mhGIPerzI4twdxaP8/0+06HBpwf345Lw=="
|
||||||
|
},
|
||||||
|
"node_modules/@types/long": {
|
||||||
|
"version": "4.0.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@types/long/-/long-4.0.2.tgz",
|
||||||
|
"integrity": "sha512-MqTGEo5bj5t157U6fA/BiDynNkn0YknVdh48CMPkTSpFTVmvao5UQmm7uEF6xBEo7qIMAlY/JSleYaE6VOdpaA=="
|
||||||
|
},
|
||||||
|
"node_modules/@types/node": {
|
||||||
|
"version": "20.14.11",
|
||||||
|
"resolved": "https://registry.npmjs.org/@types/node/-/node-20.14.11.tgz",
|
||||||
|
"integrity": "sha512-kprQpL8MMeszbz6ojB5/tU8PLN4kesnN8Gjzw349rDlNgsSzg90lAVj3llK99Dh7JON+t9AuscPPFW6mPbTnSA==",
|
||||||
|
"dependencies": {
|
||||||
|
"undici-types": "~5.26.4"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/@xenova/transformers": {
|
||||||
|
"version": "2.17.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/@xenova/transformers/-/transformers-2.17.2.tgz",
|
||||||
|
"integrity": "sha512-lZmHqzrVIkSvZdKZEx7IYY51TK0WDrC8eR0c5IMnBsO8di8are1zzw8BlLhyO2TklZKLN5UffNGs1IJwT6oOqQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"@huggingface/jinja": "^0.2.2",
|
||||||
|
"onnxruntime-web": "1.14.0",
|
||||||
|
"sharp": "^0.32.0"
|
||||||
|
},
|
||||||
|
"optionalDependencies": {
|
||||||
|
"onnxruntime-node": "1.14.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/b4a": {
|
||||||
|
"version": "1.6.6",
|
||||||
|
"resolved": "https://registry.npmjs.org/b4a/-/b4a-1.6.6.tgz",
|
||||||
|
"integrity": "sha512-5Tk1HLk6b6ctmjIkAcU/Ujv/1WqiDl0F0JdRCR80VsOcUlHcu7pWeWRlOqQLHfDEsVx9YH/aif5AG4ehoCtTmg=="
|
||||||
|
},
|
||||||
|
"node_modules/bare-events": {
|
||||||
|
"version": "2.4.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/bare-events/-/bare-events-2.4.2.tgz",
|
||||||
|
"integrity": "sha512-qMKFd2qG/36aA4GwvKq8MxnPgCQAmBWmSyLWsJcbn8v03wvIPQ/hG1Ms8bPzndZxMDoHpxez5VOS+gC9Yi24/Q==",
|
||||||
|
"optional": true
|
||||||
|
},
|
||||||
|
"node_modules/bare-fs": {
|
||||||
|
"version": "2.3.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/bare-fs/-/bare-fs-2.3.1.tgz",
|
||||||
|
"integrity": "sha512-W/Hfxc/6VehXlsgFtbB5B4xFcsCl+pAh30cYhoFyXErf6oGrwjh8SwiPAdHgpmWonKuYpZgGywN0SXt7dgsADA==",
|
||||||
|
"optional": true,
|
||||||
|
"dependencies": {
|
||||||
|
"bare-events": "^2.0.0",
|
||||||
|
"bare-path": "^2.0.0",
|
||||||
|
"bare-stream": "^2.0.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/bare-os": {
|
||||||
|
"version": "2.4.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/bare-os/-/bare-os-2.4.0.tgz",
|
||||||
|
"integrity": "sha512-v8DTT08AS/G0F9xrhyLtepoo9EJBJ85FRSMbu1pQUlAf6A8T0tEEQGMVObWeqpjhSPXsE0VGlluFBJu2fdoTNg==",
|
||||||
|
"optional": true
|
||||||
|
},
|
||||||
|
"node_modules/bare-path": {
|
||||||
|
"version": "2.1.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/bare-path/-/bare-path-2.1.3.tgz",
|
||||||
|
"integrity": "sha512-lh/eITfU8hrj9Ru5quUp0Io1kJWIk1bTjzo7JH1P5dWmQ2EL4hFUlfI8FonAhSlgIfhn63p84CDY/x+PisgcXA==",
|
||||||
|
"optional": true,
|
||||||
|
"dependencies": {
|
||||||
|
"bare-os": "^2.1.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/bare-stream": {
|
||||||
|
"version": "2.1.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/bare-stream/-/bare-stream-2.1.3.tgz",
|
||||||
|
"integrity": "sha512-tiDAH9H/kP+tvNO5sczyn9ZAA7utrSMobyDchsnyyXBuUe2FSQWbxhtuHB8jwpHYYevVo2UJpcmvvjrbHboUUQ==",
|
||||||
|
"optional": true,
|
||||||
|
"dependencies": {
|
||||||
|
"streamx": "^2.18.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/base64-js": {
|
||||||
|
"version": "1.5.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/base64-js/-/base64-js-1.5.1.tgz",
|
||||||
|
"integrity": "sha512-AKpaYlHn8t4SVbOHCy+b5+KKgvR4vrsD8vbvrbiQJps7fKDTkjkDry6ji0rUJjC0kzbNePLwzxq8iypo41qeWA==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
]
|
||||||
|
},
|
||||||
|
"node_modules/bl": {
|
||||||
|
"version": "4.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/bl/-/bl-4.1.0.tgz",
|
||||||
|
"integrity": "sha512-1W07cM9gS6DcLperZfFSj+bWLtaPGSOHWhPiGzXmvVJbRLdG82sH/Kn8EtW1VqWVA54AKf2h5k5BbnIbwF3h6w==",
|
||||||
|
"dependencies": {
|
||||||
|
"buffer": "^5.5.0",
|
||||||
|
"inherits": "^2.0.4",
|
||||||
|
"readable-stream": "^3.4.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/buffer": {
|
||||||
|
"version": "5.7.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/buffer/-/buffer-5.7.1.tgz",
|
||||||
|
"integrity": "sha512-EHcyIPBQ4BSGlvjB16k5KgAJ27CIsHY/2JBmCRReo48y9rQ3MaUzWX3KVlBa4U7MyX02HdVj0K7C3WaB3ju7FQ==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"dependencies": {
|
||||||
|
"base64-js": "^1.3.1",
|
||||||
|
"ieee754": "^1.1.13"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/chownr": {
|
||||||
|
"version": "1.1.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/chownr/-/chownr-1.1.4.tgz",
|
||||||
|
"integrity": "sha512-jJ0bqzaylmJtVnNgzTeSOs8DPavpbYgEr/b0YL8/2GO3xJEhInFmhKMUnEJQjZumK7KXGFhUy89PrsJWlakBVg=="
|
||||||
|
},
|
||||||
|
"node_modules/color": {
|
||||||
|
"version": "4.2.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/color/-/color-4.2.3.tgz",
|
||||||
|
"integrity": "sha512-1rXeuUUiGGrykh+CeBdu5Ie7OJwinCgQY0bc7GCRxy5xVHy+moaqkpL/jqQq0MtQOeYcrqEz4abc5f0KtU7W4A==",
|
||||||
|
"dependencies": {
|
||||||
|
"color-convert": "^2.0.1",
|
||||||
|
"color-string": "^1.9.0"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=12.5.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/color-convert": {
|
||||||
|
"version": "2.0.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/color-convert/-/color-convert-2.0.1.tgz",
|
||||||
|
"integrity": "sha512-RRECPsj7iu/xb5oKYcsFHSppFNnsj/52OVTRKb4zP5onXwVF3zVmmToNcOfGC+CRDpfK/U584fMg38ZHCaElKQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"color-name": "~1.1.4"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=7.0.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/color-name": {
|
||||||
|
"version": "1.1.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/color-name/-/color-name-1.1.4.tgz",
|
||||||
|
"integrity": "sha512-dOy+3AuW3a2wNbZHIuMZpTcgjGuLU/uBL/ubcZF9OXbDo8ff4O8yVp5Bf0efS8uEoYo5q4Fx7dY9OgQGXgAsQA=="
|
||||||
|
},
|
||||||
|
"node_modules/color-string": {
|
||||||
|
"version": "1.9.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/color-string/-/color-string-1.9.1.tgz",
|
||||||
|
"integrity": "sha512-shrVawQFojnZv6xM40anx4CkoDP+fZsw/ZerEMsW/pyzsRbElpsL/DBVW7q3ExxwusdNXI3lXpuhEZkzs8p5Eg==",
|
||||||
|
"dependencies": {
|
||||||
|
"color-name": "^1.0.0",
|
||||||
|
"simple-swizzle": "^0.2.2"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/decompress-response": {
|
||||||
|
"version": "6.0.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/decompress-response/-/decompress-response-6.0.0.tgz",
|
||||||
|
"integrity": "sha512-aW35yZM6Bb/4oJlZncMH2LCoZtJXTRxES17vE3hoRiowU2kWHaJKFkSBDnDR+cm9J+9QhXmREyIfv0pji9ejCQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"mimic-response": "^3.1.0"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=10"
|
||||||
|
},
|
||||||
|
"funding": {
|
||||||
|
"url": "https://github.com/sponsors/sindresorhus"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/deep-extend": {
|
||||||
|
"version": "0.6.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/deep-extend/-/deep-extend-0.6.0.tgz",
|
||||||
|
"integrity": "sha512-LOHxIOaPYdHlJRtCQfDIVZtfw/ufM8+rVj649RIHzcm/vGwQRXFt6OPqIFWsm2XEMrNIEtWR64sY1LEKD2vAOA==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=4.0.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/detect-libc": {
|
||||||
|
"version": "2.0.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/detect-libc/-/detect-libc-2.0.3.tgz",
|
||||||
|
"integrity": "sha512-bwy0MGW55bG41VqxxypOsdSdGqLwXPI/focwgTYCFMbdUiBAxLg9CFzG08sz2aqzknwiX7Hkl0bQENjg8iLByw==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=8"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/end-of-stream": {
|
||||||
|
"version": "1.4.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/end-of-stream/-/end-of-stream-1.4.4.tgz",
|
||||||
|
"integrity": "sha512-+uw1inIHVPQoaVuHzRyXd21icM+cnt4CzD5rW+NC1wjOUSTOs+Te7FOv7AhN7vS9x/oIyhLP5PR1H+phQAHu5Q==",
|
||||||
|
"dependencies": {
|
||||||
|
"once": "^1.4.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/expand-template": {
|
||||||
|
"version": "2.0.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/expand-template/-/expand-template-2.0.3.tgz",
|
||||||
|
"integrity": "sha512-XYfuKMvj4O35f/pOXLObndIRvyQ+/+6AhODh+OKWj9S9498pHHn/IMszH+gt0fBCRWMNfk1ZSp5x3AifmnI2vg==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=6"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/fast-fifo": {
|
||||||
|
"version": "1.3.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/fast-fifo/-/fast-fifo-1.3.2.tgz",
|
||||||
|
"integrity": "sha512-/d9sfos4yxzpwkDkuN7k2SqFKtYNmCTzgfEpz82x34IM9/zc8KGxQoXg1liNC/izpRM/MBdt44Nmx41ZWqk+FQ=="
|
||||||
|
},
|
||||||
|
"node_modules/flatbuffers": {
|
||||||
|
"version": "1.12.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/flatbuffers/-/flatbuffers-1.12.0.tgz",
|
||||||
|
"integrity": "sha512-c7CZADjRcl6j0PlvFy0ZqXQ67qSEZfrVPynmnL+2zPc+NtMvrF8Y0QceMo7QqnSPc7+uWjUIAbvCQ5WIKlMVdQ=="
|
||||||
|
},
|
||||||
|
"node_modules/fs-constants": {
|
||||||
|
"version": "1.0.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/fs-constants/-/fs-constants-1.0.0.tgz",
|
||||||
|
"integrity": "sha512-y6OAwoSIf7FyjMIv94u+b5rdheZEjzR63GTyZJm5qh4Bi+2YgwLCcI/fPFZkL5PSixOt6ZNKm+w+Hfp/Bciwow=="
|
||||||
|
},
|
||||||
|
"node_modules/github-from-package": {
|
||||||
|
"version": "0.0.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/github-from-package/-/github-from-package-0.0.0.tgz",
|
||||||
|
"integrity": "sha512-SyHy3T1v2NUXn29OsWdxmK6RwHD+vkj3v8en8AOBZ1wBQ/hCAQ5bAQTD02kW4W9tUp/3Qh6J8r9EvntiyCmOOw=="
|
||||||
|
},
|
||||||
|
"node_modules/guid-typescript": {
|
||||||
|
"version": "1.0.9",
|
||||||
|
"resolved": "https://registry.npmjs.org/guid-typescript/-/guid-typescript-1.0.9.tgz",
|
||||||
|
"integrity": "sha512-Y8T4vYhEfwJOTbouREvG+3XDsjr8E3kIr7uf+JZ0BYloFsttiHU0WfvANVsR7TxNUJa/WpCnw/Ino/p+DeBhBQ=="
|
||||||
|
},
|
||||||
|
"node_modules/ieee754": {
|
||||||
|
"version": "1.2.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/ieee754/-/ieee754-1.2.1.tgz",
|
||||||
|
"integrity": "sha512-dcyqhDvX1C46lXZcVqCpK+FtMRQVdIMN6/Df5js2zouUsqG7I6sFxitIC+7KYK29KdXOLHdu9zL4sFnoVQnqaA==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
]
|
||||||
|
},
|
||||||
|
"node_modules/inherits": {
|
||||||
|
"version": "2.0.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/inherits/-/inherits-2.0.4.tgz",
|
||||||
|
"integrity": "sha512-k/vGaX4/Yla3WzyMCvTQOXYeIHvqOKtnqBduzTHpzpQZzAskKMhZ2K+EnBiSM9zGSoIFeMpXKxa4dYeZIQqewQ=="
|
||||||
|
},
|
||||||
|
"node_modules/ini": {
|
||||||
|
"version": "1.3.8",
|
||||||
|
"resolved": "https://registry.npmjs.org/ini/-/ini-1.3.8.tgz",
|
||||||
|
"integrity": "sha512-JV/yugV2uzW5iMRSiZAyDtQd+nxtUnjeLt0acNdw98kKLrvuRVyB80tsREOE7yvGVgalhZ6RNXCmEHkUKBKxew=="
|
||||||
|
},
|
||||||
|
"node_modules/is-arrayish": {
|
||||||
|
"version": "0.3.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/is-arrayish/-/is-arrayish-0.3.2.tgz",
|
||||||
|
"integrity": "sha512-eVRqCvVlZbuw3GrM63ovNSNAeA1K16kaR/LRY/92w0zxQ5/1YzwblUX652i4Xs9RwAGjW9d9y6X88t8OaAJfWQ=="
|
||||||
|
},
|
||||||
|
"node_modules/long": {
|
||||||
|
"version": "4.0.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/long/-/long-4.0.0.tgz",
|
||||||
|
"integrity": "sha512-XsP+KhQif4bjX1kbuSiySJFNAehNxgLb6hPRGJ9QsUr8ajHkuXGdrHmFUTUUXhDwVX2R5bY4JNZEwbUiMhV+MA=="
|
||||||
|
},
|
||||||
|
"node_modules/mimic-response": {
|
||||||
|
"version": "3.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/mimic-response/-/mimic-response-3.1.0.tgz",
|
||||||
|
"integrity": "sha512-z0yWI+4FDrrweS8Zmt4Ej5HdJmky15+L2e6Wgn3+iK5fWzb6T3fhNFq2+MeTRb064c6Wr4N/wv0DzQTjNzHNGQ==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=10"
|
||||||
|
},
|
||||||
|
"funding": {
|
||||||
|
"url": "https://github.com/sponsors/sindresorhus"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/minimist": {
|
||||||
|
"version": "1.2.8",
|
||||||
|
"resolved": "https://registry.npmjs.org/minimist/-/minimist-1.2.8.tgz",
|
||||||
|
"integrity": "sha512-2yyAR8qBkN3YuheJanUpWC5U3bb5osDywNB8RzDVlDwDHbocAJveqqj1u8+SVD7jkWT4yvsHCpWqqWqAxb0zCA==",
|
||||||
|
"funding": {
|
||||||
|
"url": "https://github.com/sponsors/ljharb"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/mkdirp-classic": {
|
||||||
|
"version": "0.5.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/mkdirp-classic/-/mkdirp-classic-0.5.3.tgz",
|
||||||
|
"integrity": "sha512-gKLcREMhtuZRwRAfqP3RFW+TK4JqApVBtOIftVgjuABpAtpxhPGaDcfvbhNvD0B8iD1oUr/txX35NjcaY6Ns/A=="
|
||||||
|
},
|
||||||
|
"node_modules/napi-build-utils": {
|
||||||
|
"version": "1.0.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/napi-build-utils/-/napi-build-utils-1.0.2.tgz",
|
||||||
|
"integrity": "sha512-ONmRUqK7zj7DWX0D9ADe03wbwOBZxNAfF20PlGfCWQcD3+/MakShIHrMqx9YwPTfxDdF1zLeL+RGZiR9kGMLdg=="
|
||||||
|
},
|
||||||
|
"node_modules/node-abi": {
|
||||||
|
"version": "3.65.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/node-abi/-/node-abi-3.65.0.tgz",
|
||||||
|
"integrity": "sha512-ThjYBfoDNr08AWx6hGaRbfPwxKV9kVzAzOzlLKbk2CuqXE2xnCh+cbAGnwM3t8Lq4v9rUB7VfondlkBckcJrVA==",
|
||||||
|
"dependencies": {
|
||||||
|
"semver": "^7.3.5"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=10"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/node-addon-api": {
|
||||||
|
"version": "6.1.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/node-addon-api/-/node-addon-api-6.1.0.tgz",
|
||||||
|
"integrity": "sha512-+eawOlIgy680F0kBzPUNFhMZGtJ1YmqM6l4+Crf4IkImjYrO/mqPwRMh352g23uIaQKFItcQ64I7KMaJxHgAVA=="
|
||||||
|
},
|
||||||
|
"node_modules/once": {
|
||||||
|
"version": "1.4.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/once/-/once-1.4.0.tgz",
|
||||||
|
"integrity": "sha512-lNaJgI+2Q5URQBkccEKHTQOPaXdUxnZZElQTZY0MFUAuaEqe1E+Nyvgdz/aIyNi6Z9MzO5dv1H8n58/GELp3+w==",
|
||||||
|
"dependencies": {
|
||||||
|
"wrappy": "1"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/onnx-proto": {
|
||||||
|
"version": "4.0.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/onnx-proto/-/onnx-proto-4.0.4.tgz",
|
||||||
|
"integrity": "sha512-aldMOB3HRoo6q/phyB6QRQxSt895HNNw82BNyZ2CMh4bjeKv7g/c+VpAFtJuEMVfYLMbRx61hbuqnKceLeDcDA==",
|
||||||
|
"dependencies": {
|
||||||
|
"protobufjs": "^6.8.8"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/onnxruntime-common": {
|
||||||
|
"version": "1.14.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/onnxruntime-common/-/onnxruntime-common-1.14.0.tgz",
|
||||||
|
"integrity": "sha512-3LJpegM2iMNRX2wUmtYfeX/ytfOzNwAWKSq1HbRrKc9+uqG/FsEA0bbKZl1btQeZaXhC26l44NWpNUeXPII7Ew=="
|
||||||
|
},
|
||||||
|
"node_modules/onnxruntime-node": {
|
||||||
|
"version": "1.14.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/onnxruntime-node/-/onnxruntime-node-1.14.0.tgz",
|
||||||
|
"integrity": "sha512-5ba7TWomIV/9b6NH/1x/8QEeowsb+jBEvFzU6z0T4mNsFwdPqXeFUM7uxC6QeSRkEbWu3qEB0VMjrvzN/0S9+w==",
|
||||||
|
"optional": true,
|
||||||
|
"os": [
|
||||||
|
"win32",
|
||||||
|
"darwin",
|
||||||
|
"linux"
|
||||||
|
],
|
||||||
|
"dependencies": {
|
||||||
|
"onnxruntime-common": "~1.14.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/onnxruntime-web": {
|
||||||
|
"version": "1.14.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/onnxruntime-web/-/onnxruntime-web-1.14.0.tgz",
|
||||||
|
"integrity": "sha512-Kcqf43UMfW8mCydVGcX9OMXI2VN17c0p6XvR7IPSZzBf/6lteBzXHvcEVWDPmCKuGombl997HgLqj91F11DzXw==",
|
||||||
|
"dependencies": {
|
||||||
|
"flatbuffers": "^1.12.0",
|
||||||
|
"guid-typescript": "^1.0.9",
|
||||||
|
"long": "^4.0.0",
|
||||||
|
"onnx-proto": "^4.0.4",
|
||||||
|
"onnxruntime-common": "~1.14.0",
|
||||||
|
"platform": "^1.3.6"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/platform": {
|
||||||
|
"version": "1.3.6",
|
||||||
|
"resolved": "https://registry.npmjs.org/platform/-/platform-1.3.6.tgz",
|
||||||
|
"integrity": "sha512-fnWVljUchTro6RiCFvCXBbNhJc2NijN7oIQxbwsyL0buWJPG85v81ehlHI9fXrJsMNgTofEoWIQeClKpgxFLrg=="
|
||||||
|
},
|
||||||
|
"node_modules/prebuild-install": {
|
||||||
|
"version": "7.1.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/prebuild-install/-/prebuild-install-7.1.2.tgz",
|
||||||
|
"integrity": "sha512-UnNke3IQb6sgarcZIDU3gbMeTp/9SSU1DAIkil7PrqG1vZlBtY5msYccSKSHDqa3hNg436IXK+SNImReuA1wEQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"detect-libc": "^2.0.0",
|
||||||
|
"expand-template": "^2.0.3",
|
||||||
|
"github-from-package": "0.0.0",
|
||||||
|
"minimist": "^1.2.3",
|
||||||
|
"mkdirp-classic": "^0.5.3",
|
||||||
|
"napi-build-utils": "^1.0.1",
|
||||||
|
"node-abi": "^3.3.0",
|
||||||
|
"pump": "^3.0.0",
|
||||||
|
"rc": "^1.2.7",
|
||||||
|
"simple-get": "^4.0.0",
|
||||||
|
"tar-fs": "^2.0.0",
|
||||||
|
"tunnel-agent": "^0.6.0"
|
||||||
|
},
|
||||||
|
"bin": {
|
||||||
|
"prebuild-install": "bin.js"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=10"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/prebuild-install/node_modules/tar-fs": {
|
||||||
|
"version": "2.1.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/tar-fs/-/tar-fs-2.1.1.tgz",
|
||||||
|
"integrity": "sha512-V0r2Y9scmbDRLCNex/+hYzvp/zyYjvFbHPNgVTKfQvVrb6guiE/fxP+XblDNR011utopbkex2nM4dHNV6GDsng==",
|
||||||
|
"dependencies": {
|
||||||
|
"chownr": "^1.1.1",
|
||||||
|
"mkdirp-classic": "^0.5.2",
|
||||||
|
"pump": "^3.0.0",
|
||||||
|
"tar-stream": "^2.1.4"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/prebuild-install/node_modules/tar-stream": {
|
||||||
|
"version": "2.2.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/tar-stream/-/tar-stream-2.2.0.tgz",
|
||||||
|
"integrity": "sha512-ujeqbceABgwMZxEJnk2HDY2DlnUZ+9oEcb1KzTVfYHio0UE6dG71n60d8D2I4qNvleWrrXpmjpt7vZeF1LnMZQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"bl": "^4.0.3",
|
||||||
|
"end-of-stream": "^1.4.1",
|
||||||
|
"fs-constants": "^1.0.0",
|
||||||
|
"inherits": "^2.0.3",
|
||||||
|
"readable-stream": "^3.1.1"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=6"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/protobufjs": {
|
||||||
|
"version": "6.11.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/protobufjs/-/protobufjs-6.11.4.tgz",
|
||||||
|
"integrity": "sha512-5kQWPaJHi1WoCpjTGszzQ32PG2F4+wRY6BmAT4Vfw56Q2FZ4YZzK20xUYQH4YkfehY1e6QSICrJquM6xXZNcrw==",
|
||||||
|
"hasInstallScript": true,
|
||||||
|
"dependencies": {
|
||||||
|
"@protobufjs/aspromise": "^1.1.2",
|
||||||
|
"@protobufjs/base64": "^1.1.2",
|
||||||
|
"@protobufjs/codegen": "^2.0.4",
|
||||||
|
"@protobufjs/eventemitter": "^1.1.0",
|
||||||
|
"@protobufjs/fetch": "^1.1.0",
|
||||||
|
"@protobufjs/float": "^1.0.2",
|
||||||
|
"@protobufjs/inquire": "^1.1.0",
|
||||||
|
"@protobufjs/path": "^1.1.2",
|
||||||
|
"@protobufjs/pool": "^1.1.0",
|
||||||
|
"@protobufjs/utf8": "^1.1.0",
|
||||||
|
"@types/long": "^4.0.1",
|
||||||
|
"@types/node": ">=13.7.0",
|
||||||
|
"long": "^4.0.0"
|
||||||
|
},
|
||||||
|
"bin": {
|
||||||
|
"pbjs": "bin/pbjs",
|
||||||
|
"pbts": "bin/pbts"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/pump": {
|
||||||
|
"version": "3.0.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/pump/-/pump-3.0.0.tgz",
|
||||||
|
"integrity": "sha512-LwZy+p3SFs1Pytd/jYct4wpv49HiYCqd9Rlc5ZVdk0V+8Yzv6jR5Blk3TRmPL1ft69TxP0IMZGJ+WPFU2BFhww==",
|
||||||
|
"dependencies": {
|
||||||
|
"end-of-stream": "^1.1.0",
|
||||||
|
"once": "^1.3.1"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/queue-tick": {
|
||||||
|
"version": "1.0.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/queue-tick/-/queue-tick-1.0.1.tgz",
|
||||||
|
"integrity": "sha512-kJt5qhMxoszgU/62PLP1CJytzd2NKetjSRnyuj31fDd3Rlcz3fzlFdFLD1SItunPwyqEOkca6GbV612BWfaBag=="
|
||||||
|
},
|
||||||
|
"node_modules/rc": {
|
||||||
|
"version": "1.2.8",
|
||||||
|
"resolved": "https://registry.npmjs.org/rc/-/rc-1.2.8.tgz",
|
||||||
|
"integrity": "sha512-y3bGgqKj3QBdxLbLkomlohkvsA8gdAiUQlSBJnBhfn+BPxg4bc62d8TcBW15wavDfgexCgccckhcZvywyQYPOw==",
|
||||||
|
"dependencies": {
|
||||||
|
"deep-extend": "^0.6.0",
|
||||||
|
"ini": "~1.3.0",
|
||||||
|
"minimist": "^1.2.0",
|
||||||
|
"strip-json-comments": "~2.0.1"
|
||||||
|
},
|
||||||
|
"bin": {
|
||||||
|
"rc": "cli.js"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/readable-stream": {
|
||||||
|
"version": "3.6.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/readable-stream/-/readable-stream-3.6.2.tgz",
|
||||||
|
"integrity": "sha512-9u/sniCrY3D5WdsERHzHE4G2YCXqoG5FTHUiCC4SIbr6XcLZBY05ya9EKjYek9O5xOAwjGq+1JdGBAS7Q9ScoA==",
|
||||||
|
"dependencies": {
|
||||||
|
"inherits": "^2.0.3",
|
||||||
|
"string_decoder": "^1.1.1",
|
||||||
|
"util-deprecate": "^1.0.1"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">= 6"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/safe-buffer": {
|
||||||
|
"version": "5.2.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/safe-buffer/-/safe-buffer-5.2.1.tgz",
|
||||||
|
"integrity": "sha512-rp3So07KcdmmKbGvgaNxQSJr7bGVSVk5S9Eq1F+ppbRo70+YeaDxkw5Dd8NPN+GD6bjnYm2VuPuCXmpuYvmCXQ==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
]
|
||||||
|
},
|
||||||
|
"node_modules/semver": {
|
||||||
|
"version": "7.6.3",
|
||||||
|
"resolved": "https://registry.npmjs.org/semver/-/semver-7.6.3.tgz",
|
||||||
|
"integrity": "sha512-oVekP1cKtI+CTDvHWYFUcMtsK/00wmAEfyqKfNdARm8u1wNVhSgaX7A8d4UuIlUI5e84iEwOhs7ZPYRmzU9U6A==",
|
||||||
|
"bin": {
|
||||||
|
"semver": "bin/semver.js"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=10"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/sharp": {
|
||||||
|
"version": "0.32.6",
|
||||||
|
"resolved": "https://registry.npmjs.org/sharp/-/sharp-0.32.6.tgz",
|
||||||
|
"integrity": "sha512-KyLTWwgcR9Oe4d9HwCwNM2l7+J0dUQwn/yf7S0EnTtb0eVS4RxO0eUSvxPtzT4F3SY+C4K6fqdv/DO27sJ/v/w==",
|
||||||
|
"hasInstallScript": true,
|
||||||
|
"dependencies": {
|
||||||
|
"color": "^4.2.3",
|
||||||
|
"detect-libc": "^2.0.2",
|
||||||
|
"node-addon-api": "^6.1.0",
|
||||||
|
"prebuild-install": "^7.1.1",
|
||||||
|
"semver": "^7.5.4",
|
||||||
|
"simple-get": "^4.0.1",
|
||||||
|
"tar-fs": "^3.0.4",
|
||||||
|
"tunnel-agent": "^0.6.0"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": ">=14.15.0"
|
||||||
|
},
|
||||||
|
"funding": {
|
||||||
|
"url": "https://opencollective.com/libvips"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/simple-concat": {
|
||||||
|
"version": "1.0.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/simple-concat/-/simple-concat-1.0.1.tgz",
|
||||||
|
"integrity": "sha512-cSFtAPtRhljv69IK0hTVZQ+OfE9nePi/rtJmw5UjHeVyVroEqJXP1sFztKUy1qU+xvz3u/sfYJLa947b7nAN2Q==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
]
|
||||||
|
},
|
||||||
|
"node_modules/simple-get": {
|
||||||
|
"version": "4.0.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/simple-get/-/simple-get-4.0.1.tgz",
|
||||||
|
"integrity": "sha512-brv7p5WgH0jmQJr1ZDDfKDOSeWWg+OVypG99A/5vYGPqJ6pxiaHLy8nxtFjBA7oMa01ebA9gfh1uMCFqOuXxvA==",
|
||||||
|
"funding": [
|
||||||
|
{
|
||||||
|
"type": "github",
|
||||||
|
"url": "https://github.com/sponsors/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "patreon",
|
||||||
|
"url": "https://www.patreon.com/feross"
|
||||||
|
},
|
||||||
|
{
|
||||||
|
"type": "consulting",
|
||||||
|
"url": "https://feross.org/support"
|
||||||
|
}
|
||||||
|
],
|
||||||
|
"dependencies": {
|
||||||
|
"decompress-response": "^6.0.0",
|
||||||
|
"once": "^1.3.1",
|
||||||
|
"simple-concat": "^1.0.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/simple-swizzle": {
|
||||||
|
"version": "0.2.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/simple-swizzle/-/simple-swizzle-0.2.2.tgz",
|
||||||
|
"integrity": "sha512-JA//kQgZtbuY83m+xT+tXJkmJncGMTFT+C+g2h2R9uxkYIrE2yy9sgmcLhCnw57/WSD+Eh3J97FPEDFnbXnDUg==",
|
||||||
|
"dependencies": {
|
||||||
|
"is-arrayish": "^0.3.1"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/streamx": {
|
||||||
|
"version": "2.18.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/streamx/-/streamx-2.18.0.tgz",
|
||||||
|
"integrity": "sha512-LLUC1TWdjVdn1weXGcSxyTR3T4+acB6tVGXT95y0nGbca4t4o/ng1wKAGTljm9VicuCVLvRlqFYXYy5GwgM7sQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"fast-fifo": "^1.3.2",
|
||||||
|
"queue-tick": "^1.0.1",
|
||||||
|
"text-decoder": "^1.1.0"
|
||||||
|
},
|
||||||
|
"optionalDependencies": {
|
||||||
|
"bare-events": "^2.2.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/string_decoder": {
|
||||||
|
"version": "1.3.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/string_decoder/-/string_decoder-1.3.0.tgz",
|
||||||
|
"integrity": "sha512-hkRX8U1WjJFd8LsDJ2yQ/wWWxaopEsABU1XfkM8A+j0+85JAGppt16cr1Whg6KIbb4okU6Mql6BOj+uup/wKeA==",
|
||||||
|
"dependencies": {
|
||||||
|
"safe-buffer": "~5.2.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/strip-json-comments": {
|
||||||
|
"version": "2.0.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/strip-json-comments/-/strip-json-comments-2.0.1.tgz",
|
||||||
|
"integrity": "sha512-4gB8na07fecVVkOI6Rs4e7T6NOTki5EmL7TUduTs6bu3EdnSycntVJ4re8kgZA+wx9IueI2Y11bfbgwtzuE0KQ==",
|
||||||
|
"engines": {
|
||||||
|
"node": ">=0.10.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/tar-fs": {
|
||||||
|
"version": "3.0.6",
|
||||||
|
"resolved": "https://registry.npmjs.org/tar-fs/-/tar-fs-3.0.6.tgz",
|
||||||
|
"integrity": "sha512-iokBDQQkUyeXhgPYaZxmczGPhnhXZ0CmrqI+MOb/WFGS9DW5wnfrLgtjUJBvz50vQ3qfRwJ62QVoCFu8mPVu5w==",
|
||||||
|
"dependencies": {
|
||||||
|
"pump": "^3.0.0",
|
||||||
|
"tar-stream": "^3.1.5"
|
||||||
|
},
|
||||||
|
"optionalDependencies": {
|
||||||
|
"bare-fs": "^2.1.1",
|
||||||
|
"bare-path": "^2.1.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/tar-stream": {
|
||||||
|
"version": "3.1.7",
|
||||||
|
"resolved": "https://registry.npmjs.org/tar-stream/-/tar-stream-3.1.7.tgz",
|
||||||
|
"integrity": "sha512-qJj60CXt7IU1Ffyc3NJMjh6EkuCFej46zUqJ4J7pqYlThyd9bO0XBTmcOIhSzZJVWfsLks0+nle/j538YAW9RQ==",
|
||||||
|
"dependencies": {
|
||||||
|
"b4a": "^1.6.4",
|
||||||
|
"fast-fifo": "^1.2.0",
|
||||||
|
"streamx": "^2.15.0"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/text-decoder": {
|
||||||
|
"version": "1.1.1",
|
||||||
|
"resolved": "https://registry.npmjs.org/text-decoder/-/text-decoder-1.1.1.tgz",
|
||||||
|
"integrity": "sha512-8zll7REEv4GDD3x4/0pW+ppIxSNs7H1J10IKFZsuOMscumCdM2a+toDGLPA3T+1+fLBql4zbt5z83GEQGGV5VA==",
|
||||||
|
"dependencies": {
|
||||||
|
"b4a": "^1.6.4"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/tsc": {
|
||||||
|
"version": "2.0.4",
|
||||||
|
"resolved": "https://registry.npmjs.org/tsc/-/tsc-2.0.4.tgz",
|
||||||
|
"integrity": "sha512-fzoSieZI5KKJVBYGvwbVZs/J5za84f2lSTLPYf6AGiIf43tZ3GNrI1QzTLcjtyDDP4aLxd46RTZq1nQxe7+k5Q==",
|
||||||
|
"license": "MIT",
|
||||||
|
"bin": {
|
||||||
|
"tsc": "bin/tsc"
|
||||||
|
}
|
||||||
|
},
|
||||||
|
"node_modules/tunnel-agent": {
|
||||||
|
"version": "0.6.0",
|
||||||
|
"resolved": "https://registry.npmjs.org/tunnel-agent/-/tunnel-agent-0.6.0.tgz",
|
||||||
|
"integrity": "sha512-McnNiV1l8RYeY8tBgEpuodCC1mLUdbSN+CYBL7kJsJNInOP8UjDDEwdk6Mw60vdLLrr5NHKZhMAOSrR2NZuQ+w==",
|
||||||
|
"dependencies": {
|
||||||
|
"safe-buffer": "^5.0.1"
|
||||||
|
},
|
||||||
|
"engines": {
|
||||||
|
"node": "*"
|
||||||
|
}
|
||||||
|
},
|
||||||
"node_modules/typescript": {
|
"node_modules/typescript": {
|
||||||
"version": "5.5.2",
|
"version": "5.5.4",
|
||||||
"resolved": "https://registry.npmjs.org/typescript/-/typescript-5.5.2.tgz",
|
"resolved": "https://registry.npmjs.org/typescript/-/typescript-5.5.4.tgz",
|
||||||
"integrity": "sha512-NcRtPEOsPFFWjobJEtfihkLCZCXZt/os3zf8nTxjVH3RvTSxjrCamJpbExGvYOF+tFHc3pA65qpdwPbzjohhew==",
|
"integrity": "sha512-Mtq29sKDAEYP7aljRgtPOpTvOfbwRWlS6dPRzwjdE+C0R4brX/GUyhHSecbHMFLNBLcJIPt9nl9yG5TZ1weH+Q==",
|
||||||
"peer": true,
|
"dev": true,
|
||||||
|
"license": "Apache-2.0",
|
||||||
"bin": {
|
"bin": {
|
||||||
"tsc": "bin/tsc",
|
"tsc": "bin/tsc",
|
||||||
"tsserver": "bin/tsserver"
|
"tsserver": "bin/tsserver"
|
||||||
@@ -74,6 +826,21 @@
|
|||||||
"engines": {
|
"engines": {
|
||||||
"node": ">=14.17"
|
"node": ">=14.17"
|
||||||
}
|
}
|
||||||
|
},
|
||||||
|
"node_modules/undici-types": {
|
||||||
|
"version": "5.26.5",
|
||||||
|
"resolved": "https://registry.npmjs.org/undici-types/-/undici-types-5.26.5.tgz",
|
||||||
|
"integrity": "sha512-JlCMO+ehdEIKqlFxk6IfVoAUVmgz7cU7zD/h9XZ0qzeosSHmUJVOzSQvvYSYWXkFXC+IfLKSIffhv0sVZup6pA=="
|
||||||
|
},
|
||||||
|
"node_modules/util-deprecate": {
|
||||||
|
"version": "1.0.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/util-deprecate/-/util-deprecate-1.0.2.tgz",
|
||||||
|
"integrity": "sha512-EPD5q1uXyFxJpCrLnCc1nHnq3gOa6DZBocAIiI2TaSCA7VCJ1UJDMagCzIkXNsUYfD1daK//LTEQ8xiIbrHtcw=="
|
||||||
|
},
|
||||||
|
"node_modules/wrappy": {
|
||||||
|
"version": "1.0.2",
|
||||||
|
"resolved": "https://registry.npmjs.org/wrappy/-/wrappy-1.0.2.tgz",
|
||||||
|
"integrity": "sha512-l4Sp/DRseor9wL6EvV2+TuQn63dMkPjZ/sp9XkghTEbV9KlPS1xUsZ3u7/IQO4wxtcFB4bgpQPRcR3QCvezPcQ=="
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -10,9 +10,19 @@
|
|||||||
"author": "Lance Devs",
|
"author": "Lance Devs",
|
||||||
"license": "Apache-2.0",
|
"license": "Apache-2.0",
|
||||||
"dependencies": {
|
"dependencies": {
|
||||||
"@lancedb/lancedb": "file:../"
|
"@lancedb/lancedb": "file:../",
|
||||||
|
"@xenova/transformers": "^2.17.2"
|
||||||
},
|
},
|
||||||
"peerDependencies": {
|
"devDependencies": {
|
||||||
"typescript": "^5.0.0"
|
"typescript": "^5.5.4"
|
||||||
|
},
|
||||||
|
"compilerOptions": {
|
||||||
|
"target": "ESNext",
|
||||||
|
"module": "ESNext",
|
||||||
|
"moduleResolution": "Node",
|
||||||
|
"strict": true,
|
||||||
|
"esModuleInterop": true,
|
||||||
|
"skipLibCheck": true,
|
||||||
|
"forceConsistentCasingInFileNames": true
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -32,6 +32,7 @@ const _results2 = await tbl
|
|||||||
.distanceType("cosine")
|
.distanceType("cosine")
|
||||||
.limit(10)
|
.limit(10)
|
||||||
.toArray();
|
.toArray();
|
||||||
|
console.log(_results2);
|
||||||
// --8<-- [end:search2]
|
// --8<-- [end:search2]
|
||||||
|
|
||||||
console.log("search: done");
|
console.log("search: done");
|
||||||
|
|||||||
50
nodejs/examples/sentence-transformers.js
Normal file
50
nodejs/examples/sentence-transformers.js
Normal file
@@ -0,0 +1,50 @@
|
|||||||
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
|
|
||||||
|
import { LanceSchema, getRegistry } from "@lancedb/lancedb/embedding";
|
||||||
|
import { Utf8 } from "apache-arrow";
|
||||||
|
|
||||||
|
const db = await lancedb.connect("/tmp/db");
|
||||||
|
const func = await getRegistry().get("huggingface").create();
|
||||||
|
|
||||||
|
const facts = [
|
||||||
|
"Albert Einstein was a theoretical physicist.",
|
||||||
|
"The capital of France is Paris.",
|
||||||
|
"The Great Wall of China is one of the Seven Wonders of the World.",
|
||||||
|
"Python is a popular programming language.",
|
||||||
|
"Mount Everest is the highest mountain in the world.",
|
||||||
|
"Leonardo da Vinci painted the Mona Lisa.",
|
||||||
|
"Shakespeare wrote Hamlet.",
|
||||||
|
"The human body has 206 bones.",
|
||||||
|
"The speed of light is approximately 299,792 kilometers per second.",
|
||||||
|
"Water boils at 100 degrees Celsius.",
|
||||||
|
"The Earth orbits the Sun.",
|
||||||
|
"The Pyramids of Giza are located in Egypt.",
|
||||||
|
"Coffee is one of the most popular beverages in the world.",
|
||||||
|
"Tokyo is the capital city of Japan.",
|
||||||
|
"Photosynthesis is the process by which plants make their food.",
|
||||||
|
"The Pacific Ocean is the largest ocean on Earth.",
|
||||||
|
"Mozart was a prolific composer of classical music.",
|
||||||
|
"The Internet is a global network of computers.",
|
||||||
|
"Basketball is a sport played with a ball and a hoop.",
|
||||||
|
"The first computer virus was created in 1983.",
|
||||||
|
"Artificial neural networks are inspired by the human brain.",
|
||||||
|
"Deep learning is a subset of machine learning.",
|
||||||
|
"IBM's Watson won Jeopardy! in 2011.",
|
||||||
|
"The first computer programmer was Ada Lovelace.",
|
||||||
|
"The first chatbot was ELIZA, created in the 1960s.",
|
||||||
|
].map((text) => ({ text }));
|
||||||
|
|
||||||
|
const factsSchema = LanceSchema({
|
||||||
|
text: func.sourceField(new Utf8()),
|
||||||
|
vector: func.vectorField(),
|
||||||
|
});
|
||||||
|
|
||||||
|
const tbl = await db.createTable("facts", facts, {
|
||||||
|
mode: "overwrite",
|
||||||
|
schema: factsSchema,
|
||||||
|
});
|
||||||
|
|
||||||
|
const query = "How many bones are in the human body?";
|
||||||
|
const actual = await tbl.search(query).limit(1).toArray();
|
||||||
|
|
||||||
|
console.log("Answer: ", actual[0]["text"]);
|
||||||
@@ -103,50 +103,11 @@ export type IntoVector =
|
|||||||
| number[]
|
| number[]
|
||||||
| Promise<Float32Array | Float64Array | number[]>;
|
| Promise<Float32Array | Float64Array | number[]>;
|
||||||
|
|
||||||
export type FloatLike =
|
|
||||||
| import("apache-arrow-13").Float
|
|
||||||
| import("apache-arrow-14").Float
|
|
||||||
| import("apache-arrow-15").Float
|
|
||||||
| import("apache-arrow-16").Float
|
|
||||||
| import("apache-arrow-17").Float;
|
|
||||||
export type DataTypeLike =
|
|
||||||
| import("apache-arrow-13").DataType
|
|
||||||
| import("apache-arrow-14").DataType
|
|
||||||
| import("apache-arrow-15").DataType
|
|
||||||
| import("apache-arrow-16").DataType
|
|
||||||
| import("apache-arrow-17").DataType;
|
|
||||||
|
|
||||||
export function isArrowTable(value: object): value is TableLike {
|
export function isArrowTable(value: object): value is TableLike {
|
||||||
if (value instanceof ArrowTable) return true;
|
if (value instanceof ArrowTable) return true;
|
||||||
return "schema" in value && "batches" in value;
|
return "schema" in value && "batches" in value;
|
||||||
}
|
}
|
||||||
|
|
||||||
export function isDataType(value: unknown): value is DataTypeLike {
|
|
||||||
return (
|
|
||||||
value instanceof DataType ||
|
|
||||||
DataType.isNull(value) ||
|
|
||||||
DataType.isInt(value) ||
|
|
||||||
DataType.isFloat(value) ||
|
|
||||||
DataType.isBinary(value) ||
|
|
||||||
DataType.isLargeBinary(value) ||
|
|
||||||
DataType.isUtf8(value) ||
|
|
||||||
DataType.isLargeUtf8(value) ||
|
|
||||||
DataType.isBool(value) ||
|
|
||||||
DataType.isDecimal(value) ||
|
|
||||||
DataType.isDate(value) ||
|
|
||||||
DataType.isTime(value) ||
|
|
||||||
DataType.isTimestamp(value) ||
|
|
||||||
DataType.isInterval(value) ||
|
|
||||||
DataType.isDuration(value) ||
|
|
||||||
DataType.isList(value) ||
|
|
||||||
DataType.isStruct(value) ||
|
|
||||||
DataType.isUnion(value) ||
|
|
||||||
DataType.isFixedSizeBinary(value) ||
|
|
||||||
DataType.isFixedSizeList(value) ||
|
|
||||||
DataType.isMap(value) ||
|
|
||||||
DataType.isDictionary(value)
|
|
||||||
);
|
|
||||||
}
|
|
||||||
export function isNull(value: unknown): value is Null {
|
export function isNull(value: unknown): value is Null {
|
||||||
return value instanceof Null || DataType.isNull(value);
|
return value instanceof Null || DataType.isNull(value);
|
||||||
}
|
}
|
||||||
@@ -578,7 +539,7 @@ async function applyEmbeddingsFromMetadata(
|
|||||||
schema: Schema,
|
schema: Schema,
|
||||||
): Promise<ArrowTable> {
|
): Promise<ArrowTable> {
|
||||||
const registry = getRegistry();
|
const registry = getRegistry();
|
||||||
const functions = registry.parseFunctions(schema.metadata);
|
const functions = await registry.parseFunctions(schema.metadata);
|
||||||
|
|
||||||
const columns = Object.fromEntries(
|
const columns = Object.fromEntries(
|
||||||
table.schema.fields.map((field) => [
|
table.schema.fields.map((field) => [
|
||||||
|
|||||||
@@ -44,10 +44,20 @@ export interface CreateTableOptions {
|
|||||||
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
||||||
*/
|
*/
|
||||||
storageOptions?: Record<string, string>;
|
storageOptions?: Record<string, string>;
|
||||||
|
/**
|
||||||
|
* The version of the data storage format to use.
|
||||||
|
*
|
||||||
|
* The default is `legacy`, which is Lance format v1.
|
||||||
|
* `stable` is the new format, which is Lance format v2.
|
||||||
|
*/
|
||||||
|
dataStorageVersion?: string;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* If true then data files will be written with the legacy format
|
* If true then data files will be written with the legacy format
|
||||||
*
|
*
|
||||||
* The default is true while the new format is in beta
|
* The default is true while the new format is in beta
|
||||||
|
*
|
||||||
|
* Deprecated.
|
||||||
*/
|
*/
|
||||||
useLegacyFormat?: boolean;
|
useLegacyFormat?: boolean;
|
||||||
schema?: SchemaLike;
|
schema?: SchemaLike;
|
||||||
@@ -240,18 +250,26 @@ export class LocalConnection extends Connection {
|
|||||||
): Promise<Table> {
|
): Promise<Table> {
|
||||||
if (typeof nameOrOptions !== "string" && "name" in nameOrOptions) {
|
if (typeof nameOrOptions !== "string" && "name" in nameOrOptions) {
|
||||||
const { name, data, ...options } = nameOrOptions;
|
const { name, data, ...options } = nameOrOptions;
|
||||||
|
|
||||||
return this.createTable(name, data, options);
|
return this.createTable(name, data, options);
|
||||||
}
|
}
|
||||||
if (data === undefined) {
|
if (data === undefined) {
|
||||||
throw new Error("data is required");
|
throw new Error("data is required");
|
||||||
}
|
}
|
||||||
const { buf, mode } = await Table.parseTableData(data, options);
|
const { buf, mode } = await Table.parseTableData(data, options);
|
||||||
|
let dataStorageVersion = "legacy";
|
||||||
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
|
dataStorageVersion = options.useLegacyFormat ? "legacy" : "stable";
|
||||||
|
}
|
||||||
|
|
||||||
const innerTable = await this.inner.createTable(
|
const innerTable = await this.inner.createTable(
|
||||||
nameOrOptions,
|
nameOrOptions,
|
||||||
buf,
|
buf,
|
||||||
mode,
|
mode,
|
||||||
cleanseStorageOptions(options?.storageOptions),
|
cleanseStorageOptions(options?.storageOptions),
|
||||||
options?.useLegacyFormat,
|
dataStorageVersion,
|
||||||
);
|
);
|
||||||
|
|
||||||
return new LocalTable(innerTable);
|
return new LocalTable(innerTable);
|
||||||
@@ -275,6 +293,13 @@ export class LocalConnection extends Connection {
|
|||||||
metadata = registry.getTableMetadata([embeddingFunction]);
|
metadata = registry.getTableMetadata([embeddingFunction]);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
let dataStorageVersion = "legacy";
|
||||||
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
|
dataStorageVersion = options.useLegacyFormat ? "legacy" : "stable";
|
||||||
|
}
|
||||||
|
|
||||||
const table = makeEmptyTable(schema, metadata);
|
const table = makeEmptyTable(schema, metadata);
|
||||||
const buf = await fromTableToBuffer(table);
|
const buf = await fromTableToBuffer(table);
|
||||||
const innerTable = await this.inner.createEmptyTable(
|
const innerTable = await this.inner.createEmptyTable(
|
||||||
@@ -282,7 +307,7 @@ export class LocalConnection extends Connection {
|
|||||||
buf,
|
buf,
|
||||||
mode,
|
mode,
|
||||||
cleanseStorageOptions(options?.storageOptions),
|
cleanseStorageOptions(options?.storageOptions),
|
||||||
options?.useLegacyFormat,
|
dataStorageVersion,
|
||||||
);
|
);
|
||||||
return new LocalTable(innerTable);
|
return new LocalTable(innerTable);
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -15,13 +15,12 @@
|
|||||||
import "reflect-metadata";
|
import "reflect-metadata";
|
||||||
import {
|
import {
|
||||||
DataType,
|
DataType,
|
||||||
DataTypeLike,
|
|
||||||
Field,
|
Field,
|
||||||
FixedSizeList,
|
FixedSizeList,
|
||||||
|
Float,
|
||||||
Float32,
|
Float32,
|
||||||
FloatLike,
|
|
||||||
type IntoVector,
|
type IntoVector,
|
||||||
isDataType,
|
Utf8,
|
||||||
isFixedSizeList,
|
isFixedSizeList,
|
||||||
isFloat,
|
isFloat,
|
||||||
newVectorType,
|
newVectorType,
|
||||||
@@ -41,6 +40,7 @@ export interface EmbeddingFunctionConstructor<
|
|||||||
> {
|
> {
|
||||||
new (modelOptions?: T["TOptions"]): T;
|
new (modelOptions?: T["TOptions"]): T;
|
||||||
}
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* An embedding function that automatically creates vector representation for a given column.
|
* An embedding function that automatically creates vector representation for a given column.
|
||||||
*/
|
*/
|
||||||
@@ -82,6 +82,8 @@ export abstract class EmbeddingFunction<
|
|||||||
*/
|
*/
|
||||||
abstract toJSON(): Partial<M>;
|
abstract toJSON(): Partial<M>;
|
||||||
|
|
||||||
|
async init?(): Promise<void>;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* sourceField is used in combination with `LanceSchema` to provide a declarative data model
|
* sourceField is used in combination with `LanceSchema` to provide a declarative data model
|
||||||
*
|
*
|
||||||
@@ -90,11 +92,12 @@ export abstract class EmbeddingFunction<
|
|||||||
* @see {@link lancedb.LanceSchema}
|
* @see {@link lancedb.LanceSchema}
|
||||||
*/
|
*/
|
||||||
sourceField(
|
sourceField(
|
||||||
optionsOrDatatype: Partial<FieldOptions> | DataTypeLike,
|
optionsOrDatatype: Partial<FieldOptions> | DataType,
|
||||||
): [DataTypeLike, Map<string, EmbeddingFunction>] {
|
): [DataType, Map<string, EmbeddingFunction>] {
|
||||||
let datatype = isDataType(optionsOrDatatype)
|
let datatype =
|
||||||
? optionsOrDatatype
|
"datatype" in optionsOrDatatype
|
||||||
: optionsOrDatatype?.datatype;
|
? optionsOrDatatype.datatype
|
||||||
|
: optionsOrDatatype;
|
||||||
if (!datatype) {
|
if (!datatype) {
|
||||||
throw new Error("Datatype is required");
|
throw new Error("Datatype is required");
|
||||||
}
|
}
|
||||||
@@ -120,15 +123,17 @@ export abstract class EmbeddingFunction<
|
|||||||
let dims: number | undefined = this.ndims();
|
let dims: number | undefined = this.ndims();
|
||||||
|
|
||||||
// `func.vectorField(new Float32())`
|
// `func.vectorField(new Float32())`
|
||||||
if (isDataType(optionsOrDatatype)) {
|
if (optionsOrDatatype === undefined) {
|
||||||
dtype = optionsOrDatatype;
|
dtype = new Float32();
|
||||||
|
} else if (!("datatype" in optionsOrDatatype)) {
|
||||||
|
dtype = sanitizeType(optionsOrDatatype);
|
||||||
} else {
|
} else {
|
||||||
// `func.vectorField({
|
// `func.vectorField({
|
||||||
// datatype: new Float32(),
|
// datatype: new Float32(),
|
||||||
// dims: 10
|
// dims: 10
|
||||||
// })`
|
// })`
|
||||||
dims = dims ?? optionsOrDatatype?.dims;
|
dims = dims ?? optionsOrDatatype?.dims;
|
||||||
dtype = optionsOrDatatype?.datatype;
|
dtype = sanitizeType(optionsOrDatatype?.datatype);
|
||||||
}
|
}
|
||||||
|
|
||||||
if (dtype !== undefined) {
|
if (dtype !== undefined) {
|
||||||
@@ -170,7 +175,7 @@ export abstract class EmbeddingFunction<
|
|||||||
}
|
}
|
||||||
|
|
||||||
/** The datatype of the embeddings */
|
/** The datatype of the embeddings */
|
||||||
abstract embeddingDataType(): FloatLike;
|
abstract embeddingDataType(): Float;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Creates a vector representation for the given values.
|
* Creates a vector representation for the given values.
|
||||||
@@ -189,6 +194,38 @@ export abstract class EmbeddingFunction<
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* an abstract class for implementing embedding functions that take text as input
|
||||||
|
*/
|
||||||
|
export abstract class TextEmbeddingFunction<
|
||||||
|
M extends FunctionOptions = FunctionOptions,
|
||||||
|
> extends EmbeddingFunction<string, M> {
|
||||||
|
//** Generate the embeddings for the given texts */
|
||||||
|
abstract generateEmbeddings(
|
||||||
|
texts: string[],
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: we don't know what the implementor will do
|
||||||
|
...args: any[]
|
||||||
|
): Promise<number[][] | Float32Array[] | Float64Array[]>;
|
||||||
|
|
||||||
|
async computeQueryEmbeddings(data: string): Promise<Awaited<IntoVector>> {
|
||||||
|
return this.generateEmbeddings([data]).then((data) => data[0]);
|
||||||
|
}
|
||||||
|
|
||||||
|
embeddingDataType(): Float {
|
||||||
|
return new Float32();
|
||||||
|
}
|
||||||
|
|
||||||
|
override sourceField(): [DataType, Map<string, EmbeddingFunction>] {
|
||||||
|
return super.sourceField(new Utf8());
|
||||||
|
}
|
||||||
|
|
||||||
|
computeSourceEmbeddings(
|
||||||
|
data: string[],
|
||||||
|
): Promise<number[][] | Float32Array[] | Float64Array[]> {
|
||||||
|
return this.generateEmbeddings(data);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
export interface FieldOptions<T extends DataType = DataType> {
|
export interface FieldOptions<T extends DataType = DataType> {
|
||||||
datatype: T;
|
datatype: T;
|
||||||
dims?: number;
|
dims?: number;
|
||||||
|
|||||||
@@ -12,16 +12,16 @@
|
|||||||
// See the License for the specific language governing permissions and
|
// See the License for the specific language governing permissions and
|
||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
import { DataType, Field, Schema } from "../arrow";
|
import { Field, Schema } from "../arrow";
|
||||||
import { isDataType } from "../arrow";
|
|
||||||
import { sanitizeType } from "../sanitize";
|
import { sanitizeType } from "../sanitize";
|
||||||
import { EmbeddingFunction } from "./embedding_function";
|
import { EmbeddingFunction } from "./embedding_function";
|
||||||
import { EmbeddingFunctionConfig, getRegistry } from "./registry";
|
import { EmbeddingFunctionConfig, getRegistry } from "./registry";
|
||||||
|
|
||||||
export { EmbeddingFunction } from "./embedding_function";
|
export { EmbeddingFunction, TextEmbeddingFunction } from "./embedding_function";
|
||||||
|
|
||||||
// We need to explicitly export '*' so that the `register` decorator actually registers the class.
|
// We need to explicitly export '*' so that the `register` decorator actually registers the class.
|
||||||
export * from "./openai";
|
export * from "./openai";
|
||||||
|
export * from "./transformers";
|
||||||
export * from "./registry";
|
export * from "./registry";
|
||||||
|
|
||||||
/**
|
/**
|
||||||
@@ -56,15 +56,15 @@ export function LanceSchema(
|
|||||||
Partial<EmbeddingFunctionConfig>
|
Partial<EmbeddingFunctionConfig>
|
||||||
>();
|
>();
|
||||||
Object.entries(fields).forEach(([key, value]) => {
|
Object.entries(fields).forEach(([key, value]) => {
|
||||||
if (isDataType(value)) {
|
if (Array.isArray(value)) {
|
||||||
arrowFields.push(new Field(key, sanitizeType(value), true));
|
|
||||||
} else {
|
|
||||||
const [dtype, metadata] = value as [
|
const [dtype, metadata] = value as [
|
||||||
object,
|
object,
|
||||||
Map<string, EmbeddingFunction>,
|
Map<string, EmbeddingFunction>,
|
||||||
];
|
];
|
||||||
arrowFields.push(new Field(key, sanitizeType(dtype), true));
|
arrowFields.push(new Field(key, sanitizeType(dtype), true));
|
||||||
parseEmbeddingFunctions(embeddingFunctions, key, metadata);
|
parseEmbeddingFunctions(embeddingFunctions, key, metadata);
|
||||||
|
} else {
|
||||||
|
arrowFields.push(new Field(key, sanitizeType(value), true));
|
||||||
}
|
}
|
||||||
});
|
});
|
||||||
const registry = getRegistry();
|
const registry = getRegistry();
|
||||||
|
|||||||
@@ -13,7 +13,7 @@
|
|||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
import type OpenAI from "openai";
|
import type OpenAI from "openai";
|
||||||
import { type EmbeddingCreateParams } from "openai/resources";
|
import type { EmbeddingCreateParams } from "openai/resources/index";
|
||||||
import { Float, Float32 } from "../arrow";
|
import { Float, Float32 } from "../arrow";
|
||||||
import { EmbeddingFunction } from "./embedding_function";
|
import { EmbeddingFunction } from "./embedding_function";
|
||||||
import { register } from "./registry";
|
import { register } from "./registry";
|
||||||
|
|||||||
@@ -18,9 +18,14 @@ import {
|
|||||||
} from "./embedding_function";
|
} from "./embedding_function";
|
||||||
import "reflect-metadata";
|
import "reflect-metadata";
|
||||||
import { OpenAIEmbeddingFunction } from "./openai";
|
import { OpenAIEmbeddingFunction } from "./openai";
|
||||||
|
import { TransformersEmbeddingFunction } from "./transformers";
|
||||||
|
|
||||||
|
type CreateReturnType<T> = T extends { init: () => Promise<void> }
|
||||||
|
? Promise<T>
|
||||||
|
: T;
|
||||||
|
|
||||||
interface EmbeddingFunctionCreate<T extends EmbeddingFunction> {
|
interface EmbeddingFunctionCreate<T extends EmbeddingFunction> {
|
||||||
create(options?: T["TOptions"]): T;
|
create(options?: T["TOptions"]): CreateReturnType<T>;
|
||||||
}
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
@@ -32,6 +37,13 @@ interface EmbeddingFunctionCreate<T extends EmbeddingFunction> {
|
|||||||
export class EmbeddingFunctionRegistry {
|
export class EmbeddingFunctionRegistry {
|
||||||
#functions = new Map<string, EmbeddingFunctionConstructor>();
|
#functions = new Map<string, EmbeddingFunctionConstructor>();
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Get the number of registered functions
|
||||||
|
*/
|
||||||
|
length() {
|
||||||
|
return this.#functions.size;
|
||||||
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Register an embedding function
|
* Register an embedding function
|
||||||
* @param name The name of the function
|
* @param name The name of the function
|
||||||
@@ -61,38 +73,43 @@ export class EmbeddingFunctionRegistry {
|
|||||||
};
|
};
|
||||||
}
|
}
|
||||||
|
|
||||||
|
get(name: "openai"): EmbeddingFunctionCreate<OpenAIEmbeddingFunction>;
|
||||||
|
get(
|
||||||
|
name: "huggingface",
|
||||||
|
): EmbeddingFunctionCreate<TransformersEmbeddingFunction>;
|
||||||
|
get<T extends EmbeddingFunction<unknown>>(
|
||||||
|
name: string,
|
||||||
|
): EmbeddingFunctionCreate<T> | undefined;
|
||||||
/**
|
/**
|
||||||
* Fetch an embedding function by name
|
* Fetch an embedding function by name
|
||||||
* @param name The name of the function
|
* @param name The name of the function
|
||||||
*/
|
*/
|
||||||
get<T extends EmbeddingFunction<unknown>, Name extends string = "">(
|
get(name: string) {
|
||||||
name: Name extends "openai" ? "openai" : string,
|
|
||||||
//This makes it so that you can use string constants as "types", or use an explicitly supplied type
|
|
||||||
// ex:
|
|
||||||
// `registry.get("openai") -> EmbeddingFunctionCreate<OpenAIEmbeddingFunction>`
|
|
||||||
// `registry.get<MyCustomEmbeddingFunction>("my_func") -> EmbeddingFunctionCreate<MyCustomEmbeddingFunction> | undefined`
|
|
||||||
//
|
|
||||||
// the reason this is important is that we always know our built in functions are defined so the user isnt forced to do a non null/undefined
|
|
||||||
// ```ts
|
|
||||||
// const openai: OpenAIEmbeddingFunction = registry.get("openai").create()
|
|
||||||
// ```
|
|
||||||
): Name extends "openai"
|
|
||||||
? EmbeddingFunctionCreate<OpenAIEmbeddingFunction>
|
|
||||||
: EmbeddingFunctionCreate<T> | undefined {
|
|
||||||
type Output = Name extends "openai"
|
|
||||||
? EmbeddingFunctionCreate<OpenAIEmbeddingFunction>
|
|
||||||
: EmbeddingFunctionCreate<T> | undefined;
|
|
||||||
|
|
||||||
const factory = this.#functions.get(name);
|
const factory = this.#functions.get(name);
|
||||||
if (!factory) {
|
if (!factory) {
|
||||||
return undefined as Output;
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
return undefined as any;
|
||||||
|
}
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
let create: any;
|
||||||
|
if (factory.prototype.init) {
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
create = async function (options?: any) {
|
||||||
|
const instance = new factory(options);
|
||||||
|
await instance.init!();
|
||||||
|
return instance;
|
||||||
|
};
|
||||||
|
} else {
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
create = function (options?: any) {
|
||||||
|
const instance = new factory(options);
|
||||||
|
return instance;
|
||||||
|
};
|
||||||
}
|
}
|
||||||
|
|
||||||
return {
|
return {
|
||||||
create: function (options?: T["TOptions"]) {
|
create,
|
||||||
return new factory(options);
|
};
|
||||||
},
|
|
||||||
} as Output;
|
|
||||||
}
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
@@ -105,10 +122,10 @@ export class EmbeddingFunctionRegistry {
|
|||||||
/**
|
/**
|
||||||
* @ignore
|
* @ignore
|
||||||
*/
|
*/
|
||||||
parseFunctions(
|
async parseFunctions(
|
||||||
this: EmbeddingFunctionRegistry,
|
this: EmbeddingFunctionRegistry,
|
||||||
metadata: Map<string, string>,
|
metadata: Map<string, string>,
|
||||||
): Map<string, EmbeddingFunctionConfig> {
|
): Promise<Map<string, EmbeddingFunctionConfig>> {
|
||||||
if (!metadata.has("embedding_functions")) {
|
if (!metadata.has("embedding_functions")) {
|
||||||
return new Map();
|
return new Map();
|
||||||
} else {
|
} else {
|
||||||
@@ -118,25 +135,30 @@ export class EmbeddingFunctionRegistry {
|
|||||||
vectorColumn: string;
|
vectorColumn: string;
|
||||||
model: EmbeddingFunction["TOptions"];
|
model: EmbeddingFunction["TOptions"];
|
||||||
};
|
};
|
||||||
|
|
||||||
const functions = <FunctionConfig[]>(
|
const functions = <FunctionConfig[]>(
|
||||||
JSON.parse(metadata.get("embedding_functions")!)
|
JSON.parse(metadata.get("embedding_functions")!)
|
||||||
);
|
);
|
||||||
return new Map(
|
|
||||||
functions.map((f) => {
|
const items: [string, EmbeddingFunctionConfig][] = await Promise.all(
|
||||||
|
functions.map(async (f) => {
|
||||||
const fn = this.get(f.name);
|
const fn = this.get(f.name);
|
||||||
if (!fn) {
|
if (!fn) {
|
||||||
throw new Error(`Function "${f.name}" not found in registry`);
|
throw new Error(`Function "${f.name}" not found in registry`);
|
||||||
}
|
}
|
||||||
|
const func = await this.get(f.name)!.create(f.model);
|
||||||
return [
|
return [
|
||||||
f.name,
|
f.name,
|
||||||
{
|
{
|
||||||
sourceColumn: f.sourceColumn,
|
sourceColumn: f.sourceColumn,
|
||||||
vectorColumn: f.vectorColumn,
|
vectorColumn: f.vectorColumn,
|
||||||
function: this.get(f.name)!.create(f.model),
|
function: func,
|
||||||
},
|
},
|
||||||
];
|
];
|
||||||
}),
|
}),
|
||||||
);
|
);
|
||||||
|
|
||||||
|
return new Map(items);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
|||||||
193
nodejs/lancedb/embedding/transformers.ts
Normal file
193
nodejs/lancedb/embedding/transformers.ts
Normal file
@@ -0,0 +1,193 @@
|
|||||||
|
// Copyright 2023 Lance Developers.
|
||||||
|
//
|
||||||
|
// Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
|
// you may not use this file except in compliance with the License.
|
||||||
|
// You may obtain a copy of the License at
|
||||||
|
//
|
||||||
|
// http://www.apache.org/licenses/LICENSE-2.0
|
||||||
|
//
|
||||||
|
// Unless required by applicable law or agreed to in writing, software
|
||||||
|
// distributed under the License is distributed on an "AS IS" BASIS,
|
||||||
|
// WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
||||||
|
// See the License for the specific language governing permissions and
|
||||||
|
// limitations under the License.
|
||||||
|
|
||||||
|
import { Float, Float32 } from "../arrow";
|
||||||
|
import { EmbeddingFunction } from "./embedding_function";
|
||||||
|
import { register } from "./registry";
|
||||||
|
|
||||||
|
export type XenovaTransformerOptions = {
|
||||||
|
/** The wasm compatible model to use */
|
||||||
|
model: string;
|
||||||
|
/**
|
||||||
|
* The wasm compatible tokenizer to use
|
||||||
|
* If not provided, it will use the default tokenizer for the model
|
||||||
|
*/
|
||||||
|
tokenizer?: string;
|
||||||
|
/**
|
||||||
|
* The number of dimensions of the embeddings
|
||||||
|
*
|
||||||
|
* We will attempt to infer this from the model config if not provided.
|
||||||
|
* Since there isn't a standard way to get this information from the model,
|
||||||
|
* you may need to manually specify this if using a model that doesn't have a 'hidden_size' in the config.
|
||||||
|
* */
|
||||||
|
ndims?: number;
|
||||||
|
/** Options for the tokenizer */
|
||||||
|
tokenizerOptions?: {
|
||||||
|
textPair?: string | string[];
|
||||||
|
padding?: boolean | "max_length";
|
||||||
|
addSpecialTokens?: boolean;
|
||||||
|
truncation?: boolean;
|
||||||
|
maxLength?: number;
|
||||||
|
};
|
||||||
|
};
|
||||||
|
|
||||||
|
@register("huggingface")
|
||||||
|
export class TransformersEmbeddingFunction extends EmbeddingFunction<
|
||||||
|
string,
|
||||||
|
Partial<XenovaTransformerOptions>
|
||||||
|
> {
|
||||||
|
#model?: import("@xenova/transformers").PreTrainedModel;
|
||||||
|
#tokenizer?: import("@xenova/transformers").PreTrainedTokenizer;
|
||||||
|
#modelName: XenovaTransformerOptions["model"];
|
||||||
|
#initialized = false;
|
||||||
|
#tokenizerOptions: XenovaTransformerOptions["tokenizerOptions"];
|
||||||
|
#ndims?: number;
|
||||||
|
|
||||||
|
constructor(
|
||||||
|
options: Partial<XenovaTransformerOptions> = {
|
||||||
|
model: "Xenova/all-MiniLM-L6-v2",
|
||||||
|
},
|
||||||
|
) {
|
||||||
|
super();
|
||||||
|
|
||||||
|
const modelName = options?.model ?? "Xenova/all-MiniLM-L6-v2";
|
||||||
|
this.#tokenizerOptions = {
|
||||||
|
padding: true,
|
||||||
|
...options.tokenizerOptions,
|
||||||
|
};
|
||||||
|
|
||||||
|
this.#ndims = options.ndims;
|
||||||
|
this.#modelName = modelName;
|
||||||
|
}
|
||||||
|
toJSON() {
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: <explanation>
|
||||||
|
const obj: Record<string, any> = {
|
||||||
|
model: this.#modelName,
|
||||||
|
};
|
||||||
|
if (this.#ndims) {
|
||||||
|
obj["ndims"] = this.#ndims;
|
||||||
|
}
|
||||||
|
if (this.#tokenizerOptions) {
|
||||||
|
obj["tokenizerOptions"] = this.#tokenizerOptions;
|
||||||
|
}
|
||||||
|
if (this.#tokenizer) {
|
||||||
|
obj["tokenizer"] = this.#tokenizer.name;
|
||||||
|
}
|
||||||
|
return obj;
|
||||||
|
}
|
||||||
|
|
||||||
|
async init() {
|
||||||
|
let transformers;
|
||||||
|
try {
|
||||||
|
// SAFETY:
|
||||||
|
// since typescript transpiles `import` to `require`, we need to do this in an unsafe way
|
||||||
|
// We can't use `require` because `@xenova/transformers` is an ESM module
|
||||||
|
// and we can't use `import` directly because typescript will transpile it to `require`.
|
||||||
|
// and we want to remain compatible with both ESM and CJS modules
|
||||||
|
// so we use `eval` to bypass typescript for this specific import.
|
||||||
|
transformers = await eval('import("@xenova/transformers")');
|
||||||
|
} catch (e) {
|
||||||
|
throw new Error(`error loading @xenova/transformers\nReason: ${e}`);
|
||||||
|
}
|
||||||
|
|
||||||
|
try {
|
||||||
|
this.#model = await transformers.AutoModel.from_pretrained(
|
||||||
|
this.#modelName,
|
||||||
|
);
|
||||||
|
} catch (e) {
|
||||||
|
throw new Error(
|
||||||
|
`error loading model ${this.#modelName}. Make sure you are using a wasm compatible model.\nReason: ${e}`,
|
||||||
|
);
|
||||||
|
}
|
||||||
|
try {
|
||||||
|
this.#tokenizer = await transformers.AutoTokenizer.from_pretrained(
|
||||||
|
this.#modelName,
|
||||||
|
);
|
||||||
|
} catch (e) {
|
||||||
|
throw new Error(
|
||||||
|
`error loading tokenizer for ${this.#modelName}. Make sure you are using a wasm compatible model:\nReason: ${e}`,
|
||||||
|
);
|
||||||
|
}
|
||||||
|
this.#initialized = true;
|
||||||
|
}
|
||||||
|
|
||||||
|
ndims(): number {
|
||||||
|
if (this.#ndims) {
|
||||||
|
return this.#ndims;
|
||||||
|
} else {
|
||||||
|
const config = this.#model!.config;
|
||||||
|
|
||||||
|
const ndims = config["hidden_size"];
|
||||||
|
if (!ndims) {
|
||||||
|
throw new Error(
|
||||||
|
"hidden_size not found in model config, you may need to manually specify the embedding dimensions. ",
|
||||||
|
);
|
||||||
|
}
|
||||||
|
return ndims;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
embeddingDataType(): Float {
|
||||||
|
return new Float32();
|
||||||
|
}
|
||||||
|
|
||||||
|
async computeSourceEmbeddings(data: string[]): Promise<number[][]> {
|
||||||
|
// this should only happen if the user is trying to use the function directly.
|
||||||
|
// Anything going through the registry should already be initialized.
|
||||||
|
if (!this.#initialized) {
|
||||||
|
return Promise.reject(
|
||||||
|
new Error(
|
||||||
|
"something went wrong: embedding function not initialized. Please call init()",
|
||||||
|
),
|
||||||
|
);
|
||||||
|
}
|
||||||
|
const tokenizer = this.#tokenizer!;
|
||||||
|
const model = this.#model!;
|
||||||
|
|
||||||
|
const inputs = await tokenizer(data, this.#tokenizerOptions);
|
||||||
|
let tokens = await model.forward(inputs);
|
||||||
|
tokens = tokens[Object.keys(tokens)[0]];
|
||||||
|
|
||||||
|
const [nItems, nTokens] = tokens.dims;
|
||||||
|
|
||||||
|
tokens = tensorDiv(tokens.sum(1), nTokens);
|
||||||
|
|
||||||
|
// TODO: support other data types
|
||||||
|
const tokenData = tokens.data;
|
||||||
|
const stride = this.ndims();
|
||||||
|
|
||||||
|
const embeddings = [];
|
||||||
|
for (let i = 0; i < nItems; i++) {
|
||||||
|
const start = i * stride;
|
||||||
|
const end = start + stride;
|
||||||
|
const slice = tokenData.slice(start, end);
|
||||||
|
embeddings.push(Array.from(slice) as number[]); // TODO: Avoid copy here
|
||||||
|
}
|
||||||
|
return embeddings;
|
||||||
|
}
|
||||||
|
|
||||||
|
async computeQueryEmbeddings(data: string): Promise<number[]> {
|
||||||
|
return (await this.computeSourceEmbeddings([data]))[0];
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
const tensorDiv = (
|
||||||
|
src: import("@xenova/transformers").Tensor,
|
||||||
|
divBy: number,
|
||||||
|
) => {
|
||||||
|
for (let i = 0; i < src.data.length; ++i) {
|
||||||
|
src.data[i] /= divBy;
|
||||||
|
}
|
||||||
|
return src;
|
||||||
|
};
|
||||||
@@ -59,7 +59,7 @@ export {
|
|||||||
|
|
||||||
export { Index, IndexOptions, IvfPqOptions } from "./indices";
|
export { Index, IndexOptions, IvfPqOptions } from "./indices";
|
||||||
|
|
||||||
export { Table, AddDataOptions, UpdateOptions } from "./table";
|
export { Table, AddDataOptions, UpdateOptions, OptimizeOptions } from "./table";
|
||||||
|
|
||||||
export * as embedding from "./embedding";
|
export * as embedding from "./embedding";
|
||||||
|
|
||||||
|
|||||||
@@ -175,6 +175,45 @@ export class Index {
|
|||||||
static btree() {
|
static btree() {
|
||||||
return new Index(LanceDbIndex.btree());
|
return new Index(LanceDbIndex.btree());
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Create a bitmap index.
|
||||||
|
*
|
||||||
|
* A `Bitmap` index stores a bitmap for each distinct value in the column for every row.
|
||||||
|
*
|
||||||
|
* This index works best for low-cardinality columns, where the number of unique values
|
||||||
|
* is small (i.e., less than a few hundreds).
|
||||||
|
*/
|
||||||
|
static bitmap() {
|
||||||
|
return new Index(LanceDbIndex.bitmap());
|
||||||
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Create a label list index.
|
||||||
|
*
|
||||||
|
* LabelList index is a scalar index that can be used on `List<T>` columns to
|
||||||
|
* support queries with `array_contains_all` and `array_contains_any`
|
||||||
|
* using an underlying bitmap index.
|
||||||
|
*/
|
||||||
|
static labelList() {
|
||||||
|
return new Index(LanceDbIndex.labelList());
|
||||||
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Create a full text search index
|
||||||
|
*
|
||||||
|
* A full text search index is an index on a string column, so that you can conduct full
|
||||||
|
* text searches on the column.
|
||||||
|
*
|
||||||
|
* The results of a full text search are ordered by relevance measured by BM25.
|
||||||
|
*
|
||||||
|
* You can combine filters with full text search.
|
||||||
|
*
|
||||||
|
* For now, the full text search index only supports English, and doesn't support phrase search.
|
||||||
|
*/
|
||||||
|
static fts() {
|
||||||
|
return new Index(LanceDbIndex.fts());
|
||||||
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
export interface IndexOptions {
|
export interface IndexOptions {
|
||||||
|
|||||||
@@ -88,6 +88,19 @@ export interface QueryExecutionOptions {
|
|||||||
maxBatchLength?: number;
|
maxBatchLength?: number;
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Options that control the behavior of a full text search
|
||||||
|
*/
|
||||||
|
export interface FullTextSearchOptions {
|
||||||
|
/**
|
||||||
|
* The columns to search
|
||||||
|
*
|
||||||
|
* If not specified, all indexed columns will be searched.
|
||||||
|
* For now, only one column can be searched.
|
||||||
|
*/
|
||||||
|
columns?: string | string[];
|
||||||
|
}
|
||||||
|
|
||||||
/** Common methods supported by all query types */
|
/** Common methods supported by all query types */
|
||||||
export class QueryBase<NativeQueryType extends NativeQuery | NativeVectorQuery>
|
export class QueryBase<NativeQueryType extends NativeQuery | NativeVectorQuery>
|
||||||
implements AsyncIterable<RecordBatch>
|
implements AsyncIterable<RecordBatch>
|
||||||
@@ -134,6 +147,25 @@ export class QueryBase<NativeQueryType extends NativeQuery | NativeVectorQuery>
|
|||||||
return this.where(predicate);
|
return this.where(predicate);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
fullTextSearch(
|
||||||
|
query: string,
|
||||||
|
options?: Partial<FullTextSearchOptions>,
|
||||||
|
): this {
|
||||||
|
let columns: string[] | null = null;
|
||||||
|
if (options) {
|
||||||
|
if (typeof options.columns === "string") {
|
||||||
|
columns = [options.columns];
|
||||||
|
} else if (Array.isArray(options.columns)) {
|
||||||
|
columns = options.columns;
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
this.doCall((inner: NativeQueryType) =>
|
||||||
|
inner.fullTextSearch(query, columns),
|
||||||
|
);
|
||||||
|
return this;
|
||||||
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Return only the specified columns.
|
* Return only the specified columns.
|
||||||
*
|
*
|
||||||
|
|||||||
@@ -27,8 +27,7 @@ export class RestfulLanceDBClient {
|
|||||||
#apiKey: string;
|
#apiKey: string;
|
||||||
#hostOverride?: string;
|
#hostOverride?: string;
|
||||||
#closed: boolean = false;
|
#closed: boolean = false;
|
||||||
#connectionTimeout: number = 12 * 1000; // 12 seconds;
|
#timeout: number = 12 * 1000; // 12 seconds;
|
||||||
#readTimeout: number = 30 * 1000; // 30 seconds;
|
|
||||||
#session?: import("axios").AxiosInstance;
|
#session?: import("axios").AxiosInstance;
|
||||||
|
|
||||||
constructor(
|
constructor(
|
||||||
@@ -36,15 +35,13 @@ export class RestfulLanceDBClient {
|
|||||||
apiKey: string,
|
apiKey: string,
|
||||||
region: string,
|
region: string,
|
||||||
hostOverride?: string,
|
hostOverride?: string,
|
||||||
connectionTimeout?: number,
|
timeout?: number,
|
||||||
readTimeout?: number,
|
|
||||||
) {
|
) {
|
||||||
this.#dbName = dbName;
|
this.#dbName = dbName;
|
||||||
this.#apiKey = apiKey;
|
this.#apiKey = apiKey;
|
||||||
this.#region = region;
|
this.#region = region;
|
||||||
this.#hostOverride = hostOverride ?? this.#hostOverride;
|
this.#hostOverride = hostOverride ?? this.#hostOverride;
|
||||||
this.#connectionTimeout = connectionTimeout ?? this.#connectionTimeout;
|
this.#timeout = timeout ?? this.#timeout;
|
||||||
this.#readTimeout = readTimeout ?? this.#readTimeout;
|
|
||||||
}
|
}
|
||||||
|
|
||||||
// todo: cache the session.
|
// todo: cache the session.
|
||||||
@@ -59,7 +56,7 @@ export class RestfulLanceDBClient {
|
|||||||
Authorization: `Bearer ${this.#apiKey}`,
|
Authorization: `Bearer ${this.#apiKey}`,
|
||||||
},
|
},
|
||||||
transformResponse: decodeErrorData,
|
transformResponse: decodeErrorData,
|
||||||
timeout: this.#connectionTimeout,
|
timeout: this.#timeout,
|
||||||
});
|
});
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
@@ -111,7 +108,7 @@ export class RestfulLanceDBClient {
|
|||||||
params,
|
params,
|
||||||
});
|
});
|
||||||
} catch (e) {
|
} catch (e) {
|
||||||
if (e instanceof AxiosError) {
|
if (e instanceof AxiosError && e.response) {
|
||||||
response = e.response;
|
response = e.response;
|
||||||
} else {
|
} else {
|
||||||
throw e;
|
throw e;
|
||||||
@@ -165,7 +162,7 @@ export class RestfulLanceDBClient {
|
|||||||
params: new Map(Object.entries(additional.params ?? {})),
|
params: new Map(Object.entries(additional.params ?? {})),
|
||||||
});
|
});
|
||||||
} catch (e) {
|
} catch (e) {
|
||||||
if (e instanceof AxiosError) {
|
if (e instanceof AxiosError && e.response) {
|
||||||
response = e.response;
|
response = e.response;
|
||||||
} else {
|
} else {
|
||||||
throw e;
|
throw e;
|
||||||
|
|||||||
@@ -20,8 +20,7 @@ export interface RemoteConnectionOptions {
|
|||||||
apiKey?: string;
|
apiKey?: string;
|
||||||
region?: string;
|
region?: string;
|
||||||
hostOverride?: string;
|
hostOverride?: string;
|
||||||
connectionTimeout?: number;
|
timeout?: number;
|
||||||
readTimeout?: number;
|
|
||||||
}
|
}
|
||||||
|
|
||||||
export class RemoteConnection extends Connection {
|
export class RemoteConnection extends Connection {
|
||||||
@@ -33,13 +32,7 @@ export class RemoteConnection extends Connection {
|
|||||||
|
|
||||||
constructor(
|
constructor(
|
||||||
url: string,
|
url: string,
|
||||||
{
|
{ apiKey, region, hostOverride, timeout }: RemoteConnectionOptions,
|
||||||
apiKey,
|
|
||||||
region,
|
|
||||||
hostOverride,
|
|
||||||
connectionTimeout,
|
|
||||||
readTimeout,
|
|
||||||
}: RemoteConnectionOptions,
|
|
||||||
) {
|
) {
|
||||||
super();
|
super();
|
||||||
apiKey = apiKey ?? process.env.LANCEDB_API_KEY;
|
apiKey = apiKey ?? process.env.LANCEDB_API_KEY;
|
||||||
@@ -68,8 +61,7 @@ export class RemoteConnection extends Connection {
|
|||||||
this.#apiKey,
|
this.#apiKey,
|
||||||
this.#region,
|
this.#region,
|
||||||
hostOverride,
|
hostOverride,
|
||||||
connectionTimeout,
|
timeout,
|
||||||
readTimeout,
|
|
||||||
);
|
);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|||||||
@@ -340,8 +340,14 @@ export function sanitizeType(typeLike: unknown): DataType<any> {
|
|||||||
if (typeof typeLike !== "object" || typeLike === null) {
|
if (typeof typeLike !== "object" || typeLike === null) {
|
||||||
throw Error("Expected a Type but object was null/undefined");
|
throw Error("Expected a Type but object was null/undefined");
|
||||||
}
|
}
|
||||||
if (!("typeId" in typeLike) || !(typeof typeLike.typeId !== "function")) {
|
if (
|
||||||
throw Error("Expected a Type to have a typeId function");
|
!("typeId" in typeLike) ||
|
||||||
|
!(
|
||||||
|
typeof typeLike.typeId !== "function" ||
|
||||||
|
typeof typeLike.typeId !== "number"
|
||||||
|
)
|
||||||
|
) {
|
||||||
|
throw Error("Expected a Type to have a typeId property");
|
||||||
}
|
}
|
||||||
let typeId: Type;
|
let typeId: Type;
|
||||||
if (typeof typeLike.typeId === "function") {
|
if (typeof typeLike.typeId === "function") {
|
||||||
|
|||||||
@@ -270,19 +270,23 @@ export abstract class Table {
|
|||||||
* @returns {Query} A builder that can be used to parameterize the query
|
* @returns {Query} A builder that can be used to parameterize the query
|
||||||
*/
|
*/
|
||||||
abstract query(): Query;
|
abstract query(): Query;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Create a search query to find the nearest neighbors
|
* Create a search query to find the nearest neighbors
|
||||||
* of the given query vector
|
* of the given query
|
||||||
* @param {string} query - the query. This will be converted to a vector using the table's provided embedding function
|
* @param {string | IntoVector} query - the query, a vector or string
|
||||||
* @note If no embedding functions are defined in the table, this will error when collecting the results.
|
* @param {string} queryType - the type of the query, "vector", "fts", or "auto"
|
||||||
|
* @param {string | string[]} ftsColumns - the columns to search in for full text search
|
||||||
|
* for now, only one column can be searched at a time.
|
||||||
|
*
|
||||||
|
* when "auto" is used, if the query is a string and an embedding function is defined, it will be treated as a vector query
|
||||||
|
* if the query is a string and no embedding function is defined, it will be treated as a full text search query
|
||||||
*/
|
*/
|
||||||
abstract search(query: string): VectorQuery;
|
abstract search(
|
||||||
/**
|
query: string | IntoVector,
|
||||||
* Create a search query to find the nearest neighbors
|
queryType?: string,
|
||||||
* of the given query vector
|
ftsColumns?: string | string[],
|
||||||
* @param {IntoVector} query - the query vector
|
): VectorQuery | Query;
|
||||||
*/
|
|
||||||
abstract search(query: IntoVector): VectorQuery;
|
|
||||||
/**
|
/**
|
||||||
* Search the table with a given query vector.
|
* Search the table with a given query vector.
|
||||||
*
|
*
|
||||||
@@ -490,7 +494,7 @@ export class LocalTable extends Table {
|
|||||||
const mode = options?.mode ?? "append";
|
const mode = options?.mode ?? "append";
|
||||||
const schema = await this.schema();
|
const schema = await this.schema();
|
||||||
const registry = getRegistry();
|
const registry = getRegistry();
|
||||||
const functions = registry.parseFunctions(schema.metadata);
|
const functions = await registry.parseFunctions(schema.metadata);
|
||||||
|
|
||||||
const buffer = await fromDataToBuffer(
|
const buffer = await fromDataToBuffer(
|
||||||
data,
|
data,
|
||||||
@@ -578,10 +582,34 @@ export class LocalTable extends Table {
|
|||||||
query(): Query {
|
query(): Query {
|
||||||
return new Query(this.inner);
|
return new Query(this.inner);
|
||||||
}
|
}
|
||||||
search(query: string | IntoVector): VectorQuery {
|
|
||||||
|
search(
|
||||||
|
query: string | IntoVector,
|
||||||
|
queryType: string = "auto",
|
||||||
|
ftsColumns?: string | string[],
|
||||||
|
): VectorQuery | Query {
|
||||||
if (typeof query !== "string") {
|
if (typeof query !== "string") {
|
||||||
|
if (queryType === "fts") {
|
||||||
|
throw new Error("Cannot perform full text search on a vector query");
|
||||||
|
}
|
||||||
return this.vectorSearch(query);
|
return this.vectorSearch(query);
|
||||||
} else {
|
}
|
||||||
|
|
||||||
|
// If the query is a string, we need to determine if it is a vector query or a full text search query
|
||||||
|
if (queryType === "fts") {
|
||||||
|
return this.query().fullTextSearch(query, {
|
||||||
|
columns: ftsColumns,
|
||||||
|
});
|
||||||
|
}
|
||||||
|
|
||||||
|
// The query type is auto or vector
|
||||||
|
// fall back to full text search if no embedding functions are defined and the query is a string
|
||||||
|
if (queryType === "auto" && getRegistry().length() === 0) {
|
||||||
|
return this.query().fullTextSearch(query, {
|
||||||
|
columns: ftsColumns,
|
||||||
|
});
|
||||||
|
}
|
||||||
|
|
||||||
const queryPromise = this.getEmbeddingFunctions().then(
|
const queryPromise = this.getEmbeddingFunctions().then(
|
||||||
async (functions) => {
|
async (functions) => {
|
||||||
// TODO: Support multiple embedding functions
|
// TODO: Support multiple embedding functions
|
||||||
@@ -599,7 +627,6 @@ export class LocalTable extends Table {
|
|||||||
|
|
||||||
return this.query().nearestTo(queryPromise);
|
return this.query().nearestTo(queryPromise);
|
||||||
}
|
}
|
||||||
}
|
|
||||||
|
|
||||||
vectorSearch(vector: IntoVector): VectorQuery {
|
vectorSearch(vector: IntoVector): VectorQuery {
|
||||||
return this.query().nearestTo(vector);
|
return this.query().nearestTo(vector);
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-arm64",
|
"name": "@lancedb/lancedb-darwin-arm64",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.darwin-arm64.node",
|
"main": "lancedb.darwin-arm64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-x64",
|
"name": "@lancedb/lancedb-darwin-x64",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.darwin-x64.node",
|
"main": "lancedb.darwin-x64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.linux-arm64-gnu.node",
|
"main": "lancedb.linux-arm64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-x64-gnu",
|
"name": "@lancedb/lancedb-linux-x64-gnu",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.linux-x64-gnu.node",
|
"main": "lancedb.linux-x64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-win32-x64-msvc",
|
"name": "@lancedb/lancedb-win32-x64-msvc",
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"os": ["win32"],
|
"os": ["win32"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.win32-x64-msvc.node",
|
"main": "lancedb.win32-x64-msvc.node",
|
||||||
|
|||||||
771
nodejs/package-lock.json
generated
771
nodejs/package-lock.json
generated
File diff suppressed because it is too large
Load Diff
@@ -10,7 +10,7 @@
|
|||||||
"vector database",
|
"vector database",
|
||||||
"ann"
|
"ann"
|
||||||
],
|
],
|
||||||
"version": "0.7.1",
|
"version": "0.9.0",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"exports": {
|
"exports": {
|
||||||
".": "./dist/index.js",
|
".": "./dist/index.js",
|
||||||
@@ -32,12 +32,13 @@
|
|||||||
},
|
},
|
||||||
"license": "Apache 2.0",
|
"license": "Apache 2.0",
|
||||||
"devDependencies": {
|
"devDependencies": {
|
||||||
|
"@aws-sdk/client-dynamodb": "^3.33.0",
|
||||||
"@aws-sdk/client-kms": "^3.33.0",
|
"@aws-sdk/client-kms": "^3.33.0",
|
||||||
"@aws-sdk/client-s3": "^3.33.0",
|
"@aws-sdk/client-s3": "^3.33.0",
|
||||||
"@aws-sdk/client-dynamodb": "^3.33.0",
|
|
||||||
"@biomejs/biome": "^1.7.3",
|
"@biomejs/biome": "^1.7.3",
|
||||||
"@jest/globals": "^29.7.0",
|
"@jest/globals": "^29.7.0",
|
||||||
"@napi-rs/cli": "^2.18.3",
|
"@napi-rs/cli": "^2.18.3",
|
||||||
|
"@types/axios": "^0.14.0",
|
||||||
"@types/jest": "^29.1.2",
|
"@types/jest": "^29.1.2",
|
||||||
"@types/tmp": "^0.2.6",
|
"@types/tmp": "^0.2.6",
|
||||||
"apache-arrow-13": "npm:apache-arrow@13.0.0",
|
"apache-arrow-13": "npm:apache-arrow@13.0.0",
|
||||||
@@ -52,9 +53,8 @@
|
|||||||
"ts-jest": "^29.1.2",
|
"ts-jest": "^29.1.2",
|
||||||
"typedoc": "^0.26.4",
|
"typedoc": "^0.26.4",
|
||||||
"typedoc-plugin-markdown": "^4.2.1",
|
"typedoc-plugin-markdown": "^4.2.1",
|
||||||
"typescript": "^5.3.3",
|
"typescript": "^5.5.4",
|
||||||
"typescript-eslint": "^7.1.0",
|
"typescript-eslint": "^7.1.0"
|
||||||
"@types/axios": "^0.14.0"
|
|
||||||
},
|
},
|
||||||
"ava": {
|
"ava": {
|
||||||
"timeout": "3m"
|
"timeout": "3m"
|
||||||
@@ -85,6 +85,7 @@
|
|||||||
"reflect-metadata": "^0.2.2"
|
"reflect-metadata": "^0.2.2"
|
||||||
},
|
},
|
||||||
"optionalDependencies": {
|
"optionalDependencies": {
|
||||||
|
"@xenova/transformers": ">=2.17 < 3",
|
||||||
"openai": "^4.29.2"
|
"openai": "^4.29.2"
|
||||||
},
|
},
|
||||||
"peerDependencies": {
|
"peerDependencies": {
|
||||||
|
|||||||
@@ -13,13 +13,16 @@
|
|||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
use std::collections::HashMap;
|
use std::collections::HashMap;
|
||||||
|
use std::str::FromStr;
|
||||||
|
|
||||||
use napi::bindgen_prelude::*;
|
use napi::bindgen_prelude::*;
|
||||||
use napi_derive::*;
|
use napi_derive::*;
|
||||||
|
|
||||||
use crate::table::Table;
|
use crate::table::Table;
|
||||||
use crate::ConnectionOptions;
|
use crate::ConnectionOptions;
|
||||||
use lancedb::connection::{ConnectBuilder, Connection as LanceDBConnection, CreateTableMode};
|
use lancedb::connection::{
|
||||||
|
ConnectBuilder, Connection as LanceDBConnection, CreateTableMode, LanceFileVersion,
|
||||||
|
};
|
||||||
use lancedb::ipc::{ipc_file_to_batches, ipc_file_to_schema};
|
use lancedb::ipc::{ipc_file_to_batches, ipc_file_to_schema};
|
||||||
|
|
||||||
#[napi]
|
#[napi]
|
||||||
@@ -120,7 +123,7 @@ impl Connection {
|
|||||||
buf: Buffer,
|
buf: Buffer,
|
||||||
mode: String,
|
mode: String,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_options: Option<String>,
|
||||||
) -> napi::Result<Table> {
|
) -> napi::Result<Table> {
|
||||||
let batches = ipc_file_to_batches(buf.to_vec())
|
let batches = ipc_file_to_batches(buf.to_vec())
|
||||||
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
||||||
@@ -131,8 +134,11 @@ impl Connection {
|
|||||||
builder = builder.storage_option(key, value);
|
builder = builder.storage_option(key, value);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_option) = data_storage_options.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_option)
|
||||||
|
.map_err(|e| napi::Error::from_reason(format!("{}", e)))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
let tbl = builder
|
let tbl = builder
|
||||||
.execute()
|
.execute()
|
||||||
@@ -148,7 +154,7 @@ impl Connection {
|
|||||||
schema_buf: Buffer,
|
schema_buf: Buffer,
|
||||||
mode: String,
|
mode: String,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_options: Option<String>,
|
||||||
) -> napi::Result<Table> {
|
) -> napi::Result<Table> {
|
||||||
let schema = ipc_file_to_schema(schema_buf.to_vec()).map_err(|e| {
|
let schema = ipc_file_to_schema(schema_buf.to_vec()).map_err(|e| {
|
||||||
napi::Error::from_reason(format!("Failed to marshal schema from JS to Rust: {}", e))
|
napi::Error::from_reason(format!("Failed to marshal schema from JS to Rust: {}", e))
|
||||||
@@ -163,8 +169,11 @@ impl Connection {
|
|||||||
builder = builder.storage_option(key, value);
|
builder = builder.storage_option(key, value);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_option) = data_storage_options.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_option)
|
||||||
|
.map_err(|e| napi::Error::from_reason(format!("{}", e)))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
let tbl = builder
|
let tbl = builder
|
||||||
.execute()
|
.execute()
|
||||||
|
|||||||
@@ -14,7 +14,7 @@
|
|||||||
|
|
||||||
use std::sync::Mutex;
|
use std::sync::Mutex;
|
||||||
|
|
||||||
use lancedb::index::scalar::BTreeIndexBuilder;
|
use lancedb::index::scalar::{BTreeIndexBuilder, FtsIndexBuilder};
|
||||||
use lancedb::index::vector::IvfPqIndexBuilder;
|
use lancedb::index::vector::IvfPqIndexBuilder;
|
||||||
use lancedb::index::Index as LanceDbIndex;
|
use lancedb::index::Index as LanceDbIndex;
|
||||||
use napi_derive::napi;
|
use napi_derive::napi;
|
||||||
@@ -76,4 +76,25 @@ impl Index {
|
|||||||
inner: Mutex::new(Some(LanceDbIndex::BTree(BTreeIndexBuilder::default()))),
|
inner: Mutex::new(Some(LanceDbIndex::BTree(BTreeIndexBuilder::default()))),
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[napi(factory)]
|
||||||
|
pub fn bitmap() -> Self {
|
||||||
|
Self {
|
||||||
|
inner: Mutex::new(Some(LanceDbIndex::Bitmap(Default::default()))),
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#[napi(factory)]
|
||||||
|
pub fn label_list() -> Self {
|
||||||
|
Self {
|
||||||
|
inner: Mutex::new(Some(LanceDbIndex::LabelList(Default::default()))),
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#[napi(factory)]
|
||||||
|
pub fn fts() -> Self {
|
||||||
|
Self {
|
||||||
|
inner: Mutex::new(Some(LanceDbIndex::FTS(FtsIndexBuilder::default()))),
|
||||||
|
}
|
||||||
|
}
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -12,6 +12,7 @@
|
|||||||
// See the License for the specific language governing permissions and
|
// See the License for the specific language governing permissions and
|
||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
|
use lancedb::index::scalar::FullTextSearchQuery;
|
||||||
use lancedb::query::ExecutableQuery;
|
use lancedb::query::ExecutableQuery;
|
||||||
use lancedb::query::Query as LanceDbQuery;
|
use lancedb::query::Query as LanceDbQuery;
|
||||||
use lancedb::query::QueryBase;
|
use lancedb::query::QueryBase;
|
||||||
@@ -42,6 +43,12 @@ impl Query {
|
|||||||
self.inner = self.inner.clone().only_if(predicate);
|
self.inner = self.inner.clone().only_if(predicate);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[napi]
|
||||||
|
pub fn full_text_search(&mut self, query: String, columns: Option<Vec<String>>) {
|
||||||
|
let query = FullTextSearchQuery::new(query).columns(columns);
|
||||||
|
self.inner = self.inner.clone().full_text_search(query);
|
||||||
|
}
|
||||||
|
|
||||||
#[napi]
|
#[napi]
|
||||||
pub fn select(&mut self, columns: Vec<(String, String)>) {
|
pub fn select(&mut self, columns: Vec<(String, String)>) {
|
||||||
self.inner = self.inner.clone().select(Select::dynamic(&columns));
|
self.inner = self.inner.clone().select(Select::dynamic(&columns));
|
||||||
@@ -138,6 +145,12 @@ impl VectorQuery {
|
|||||||
self.inner = self.inner.clone().only_if(predicate);
|
self.inner = self.inner.clone().only_if(predicate);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[napi]
|
||||||
|
pub fn full_text_search(&mut self, query: String, columns: Option<Vec<String>>) {
|
||||||
|
let query = FullTextSearchQuery::new(query).columns(columns);
|
||||||
|
self.inner = self.inner.clone().full_text_search(query);
|
||||||
|
}
|
||||||
|
|
||||||
#[napi]
|
#[napi]
|
||||||
pub fn select(&mut self, columns: Vec<(String, String)>) {
|
pub fn select(&mut self, columns: Vec<(String, String)>) {
|
||||||
self.inner = self.inner.clone().select(Select::dynamic(&columns));
|
self.inner = self.inner.clone().select(Select::dynamic(&columns));
|
||||||
|
|||||||
@@ -293,6 +293,7 @@ impl Table {
|
|||||||
.optimize(OptimizeAction::Prune {
|
.optimize(OptimizeAction::Prune {
|
||||||
older_than,
|
older_than,
|
||||||
delete_unverified: None,
|
delete_unverified: None,
|
||||||
|
error_if_tagged_old_versions: None,
|
||||||
})
|
})
|
||||||
.await
|
.await
|
||||||
.default_error()?
|
.default_error()?
|
||||||
|
|||||||
@@ -9,7 +9,8 @@
|
|||||||
"allowJs": true,
|
"allowJs": true,
|
||||||
"resolveJsonModule": true,
|
"resolveJsonModule": true,
|
||||||
"emitDecoratorMetadata": true,
|
"emitDecoratorMetadata": true,
|
||||||
"experimentalDecorators": true
|
"experimentalDecorators": true,
|
||||||
|
"moduleResolution": "Node"
|
||||||
},
|
},
|
||||||
"exclude": ["./dist/*"],
|
"exclude": ["./dist/*"],
|
||||||
"typedocOptions": {
|
"typedocOptions": {
|
||||||
|
|||||||
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.10.2"
|
current_version = "0.13.0-beta.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb-python"
|
name = "lancedb-python"
|
||||||
version = "0.10.2"
|
version = "0.13.0-beta.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
description = "Python bindings for LanceDB"
|
description = "Python bindings for LanceDB"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
@@ -14,11 +14,13 @@ name = "_lancedb"
|
|||||||
crate-type = ["cdylib"]
|
crate-type = ["cdylib"]
|
||||||
|
|
||||||
[dependencies]
|
[dependencies]
|
||||||
arrow = { version = "51.0.0", features = ["pyarrow"] }
|
arrow = { version = "52.1", features = ["pyarrow"] }
|
||||||
lancedb = { path = "../rust/lancedb" }
|
lancedb = { path = "../rust/lancedb" }
|
||||||
env_logger = "0.10"
|
env_logger = "0.10"
|
||||||
pyo3 = { version = "0.20", features = ["extension-module", "abi3-py38"] }
|
pyo3 = { version = "0.21", features = ["extension-module", "abi3-py38", "gil-refs"] }
|
||||||
pyo3-asyncio = { version = "0.20", features = ["attributes", "tokio-runtime"] }
|
# Using this fork for now: https://github.com/awestlake87/pyo3-asyncio/issues/119
|
||||||
|
# pyo3-asyncio = { version = "0.20", features = ["attributes", "tokio-runtime"] }
|
||||||
|
pyo3-asyncio-0-21 = { version = "0.21.0", features = ["attributes", "tokio-runtime"] }
|
||||||
|
|
||||||
# Prevent dynamic linking of lzma, which comes from datafusion
|
# Prevent dynamic linking of lzma, which comes from datafusion
|
||||||
lzma-sys = { version = "*", features = ["static"] }
|
lzma-sys = { version = "*", features = ["static"] }
|
||||||
|
|||||||
@@ -3,7 +3,7 @@ name = "lancedb"
|
|||||||
# version in Cargo.toml
|
# version in Cargo.toml
|
||||||
dependencies = [
|
dependencies = [
|
||||||
"deprecation",
|
"deprecation",
|
||||||
"pylance==0.14.1",
|
"pylance==0.16.1",
|
||||||
"ratelimiter~=1.0",
|
"ratelimiter~=1.0",
|
||||||
"requests>=2.31.0",
|
"requests>=2.31.0",
|
||||||
"retry>=0.9.2",
|
"retry>=0.9.2",
|
||||||
@@ -56,7 +56,7 @@ tests = [
|
|||||||
"pytest-asyncio",
|
"pytest-asyncio",
|
||||||
"duckdb",
|
"duckdb",
|
||||||
"pytz",
|
"pytz",
|
||||||
"polars>=0.19",
|
"polars>=0.19, <=1.3.0",
|
||||||
"tantivy",
|
"tantivy",
|
||||||
]
|
]
|
||||||
dev = ["ruff", "pre-commit"]
|
dev = ["ruff", "pre-commit"]
|
||||||
@@ -76,6 +76,7 @@ embeddings = [
|
|||||||
"awscli>=1.29.57",
|
"awscli>=1.29.57",
|
||||||
"botocore>=1.31.57",
|
"botocore>=1.31.57",
|
||||||
"ollama",
|
"ollama",
|
||||||
|
"ibm-watsonx-ai>=1.1.2",
|
||||||
]
|
]
|
||||||
azure = ["adlfs>=2024.2.0"]
|
azure = ["adlfs>=2024.2.0"]
|
||||||
|
|
||||||
|
|||||||
@@ -24,7 +24,7 @@ class Connection(object):
|
|||||||
mode: str,
|
mode: str,
|
||||||
data: pa.RecordBatchReader,
|
data: pa.RecordBatchReader,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
data_storage_version: Optional[str] = None,
|
||||||
) -> Table: ...
|
) -> Table: ...
|
||||||
async def create_empty_table(
|
async def create_empty_table(
|
||||||
self,
|
self,
|
||||||
@@ -32,7 +32,7 @@ class Connection(object):
|
|||||||
mode: str,
|
mode: str,
|
||||||
schema: pa.Schema,
|
schema: pa.Schema,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
data_storage_version: Optional[str] = None,
|
||||||
) -> Table: ...
|
) -> Table: ...
|
||||||
|
|
||||||
class Table:
|
class Table:
|
||||||
|
|||||||
@@ -560,6 +560,7 @@ class AsyncConnection(object):
|
|||||||
fill_value: Optional[float] = None,
|
fill_value: Optional[float] = None,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
*,
|
*,
|
||||||
|
data_storage_version: Optional[str] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
use_legacy_format: Optional[bool] = None,
|
||||||
) -> AsyncTable:
|
) -> AsyncTable:
|
||||||
"""Create an [AsyncTable][lancedb.table.AsyncTable] in the database.
|
"""Create an [AsyncTable][lancedb.table.AsyncTable] in the database.
|
||||||
@@ -603,9 +604,15 @@ class AsyncConnection(object):
|
|||||||
connection will be inherited by the table, but can be overridden here.
|
connection will be inherited by the table, but can be overridden here.
|
||||||
See available options at
|
See available options at
|
||||||
https://lancedb.github.io/lancedb/guides/storage/
|
https://lancedb.github.io/lancedb/guides/storage/
|
||||||
use_legacy_format: bool, optional, default True
|
data_storage_version: optional, str, default "legacy"
|
||||||
|
The version of the data storage format to use. Newer versions are more
|
||||||
|
efficient but require newer versions of lance to read. The default is
|
||||||
|
"legacy" which will use the legacy v1 version. See the user guide
|
||||||
|
for more details.
|
||||||
|
use_legacy_format: bool, optional, default True. (Deprecated)
|
||||||
If True, use the legacy format for the table. If False, use the new format.
|
If True, use the legacy format for the table. If False, use the new format.
|
||||||
The default is True while the new format is in beta.
|
The default is True while the new format is in beta.
|
||||||
|
This method is deprecated, use `data_storage_version` instead.
|
||||||
|
|
||||||
|
|
||||||
Returns
|
Returns
|
||||||
@@ -732,7 +739,7 @@ class AsyncConnection(object):
|
|||||||
fill_value = 0.0
|
fill_value = 0.0
|
||||||
|
|
||||||
if data is not None:
|
if data is not None:
|
||||||
data = _sanitize_data(
|
data, schema = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
schema,
|
schema,
|
||||||
metadata=metadata,
|
metadata=metadata,
|
||||||
@@ -765,13 +772,18 @@ class AsyncConnection(object):
|
|||||||
if mode == "create" and exist_ok:
|
if mode == "create" and exist_ok:
|
||||||
mode = "exist_ok"
|
mode = "exist_ok"
|
||||||
|
|
||||||
|
if not data_storage_version:
|
||||||
|
data_storage_version = (
|
||||||
|
"legacy" if use_legacy_format is None or use_legacy_format else "stable"
|
||||||
|
)
|
||||||
|
|
||||||
if data is None:
|
if data is None:
|
||||||
new_table = await self._inner.create_empty_table(
|
new_table = await self._inner.create_empty_table(
|
||||||
name,
|
name,
|
||||||
mode,
|
mode,
|
||||||
schema,
|
schema,
|
||||||
storage_options=storage_options,
|
storage_options=storage_options,
|
||||||
use_legacy_format=use_legacy_format,
|
data_storage_version=data_storage_version,
|
||||||
)
|
)
|
||||||
else:
|
else:
|
||||||
data = data_to_reader(data, schema)
|
data = data_to_reader(data, schema)
|
||||||
@@ -780,7 +792,7 @@ class AsyncConnection(object):
|
|||||||
mode,
|
mode,
|
||||||
data,
|
data,
|
||||||
storage_options=storage_options,
|
storage_options=storage_options,
|
||||||
use_legacy_format=use_legacy_format,
|
data_storage_version=data_storage_version,
|
||||||
)
|
)
|
||||||
|
|
||||||
return AsyncTable(new_table)
|
return AsyncTable(new_table)
|
||||||
|
|||||||
@@ -26,3 +26,4 @@ from .transformers import TransformersEmbeddingFunction, ColbertEmbeddings
|
|||||||
from .imagebind import ImageBindEmbeddings
|
from .imagebind import ImageBindEmbeddings
|
||||||
from .utils import with_embeddings
|
from .utils import with_embeddings
|
||||||
from .jinaai import JinaEmbeddings
|
from .jinaai import JinaEmbeddings
|
||||||
|
from .watsonx import WatsonxEmbeddings
|
||||||
|
|||||||
111
python/python/lancedb/embeddings/watsonx.py
Normal file
111
python/python/lancedb/embeddings/watsonx.py
Normal file
@@ -0,0 +1,111 @@
|
|||||||
|
# Copyright (c) 2023. LanceDB Developers
|
||||||
|
#
|
||||||
|
# Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
|
# you may not use this file except in compliance with the License.
|
||||||
|
# You may obtain a copy of the License at
|
||||||
|
# http://www.apache.org/licenses/LICENSE-2.0
|
||||||
|
#
|
||||||
|
# Unless required by applicable law or agreed to in writing, software
|
||||||
|
# distributed under the License is distributed on an "AS IS" BASIS,
|
||||||
|
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
||||||
|
# See the License for the specific language governing permissions and
|
||||||
|
# limitations under the License.
|
||||||
|
import os
|
||||||
|
from functools import cached_property
|
||||||
|
from typing import List, Optional, Dict, Union
|
||||||
|
|
||||||
|
from ..util import attempt_import_or_raise
|
||||||
|
from .base import TextEmbeddingFunction
|
||||||
|
from .registry import register
|
||||||
|
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
DEFAULT_WATSONX_URL = "https://us-south.ml.cloud.ibm.com"
|
||||||
|
|
||||||
|
MODELS_DIMS = {
|
||||||
|
"ibm/slate-125m-english-rtrvr": 768,
|
||||||
|
"ibm/slate-30m-english-rtrvr": 384,
|
||||||
|
"sentence-transformers/all-minilm-l12-v2": 384,
|
||||||
|
"intfloat/multilingual-e5-large": 1024,
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
@register("watsonx")
|
||||||
|
class WatsonxEmbeddings(TextEmbeddingFunction):
|
||||||
|
"""
|
||||||
|
API Docs:
|
||||||
|
---------
|
||||||
|
https://cloud.ibm.com/apidocs/watsonx-ai#text-embeddings
|
||||||
|
|
||||||
|
Supported embedding models:
|
||||||
|
---------------------------
|
||||||
|
https://dataplatform.cloud.ibm.com/docs/content/wsj/analyze-data/fm-models-embed.html?context=wx
|
||||||
|
"""
|
||||||
|
|
||||||
|
name: str = "ibm/slate-125m-english-rtrvr"
|
||||||
|
api_key: Optional[str] = None
|
||||||
|
project_id: Optional[str] = None
|
||||||
|
url: Optional[str] = None
|
||||||
|
params: Optional[Dict] = None
|
||||||
|
|
||||||
|
@staticmethod
|
||||||
|
def model_names():
|
||||||
|
return [
|
||||||
|
"ibm/slate-125m-english-rtrvr",
|
||||||
|
"ibm/slate-30m-english-rtrvr",
|
||||||
|
"sentence-transformers/all-minilm-l12-v2",
|
||||||
|
"intfloat/multilingual-e5-large",
|
||||||
|
]
|
||||||
|
|
||||||
|
def ndims(self):
|
||||||
|
return self._ndims
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def _ndims(self):
|
||||||
|
if self.name not in MODELS_DIMS:
|
||||||
|
raise ValueError(f"Unknown model name {self.name}")
|
||||||
|
return MODELS_DIMS[self.name]
|
||||||
|
|
||||||
|
def generate_embeddings(
|
||||||
|
self,
|
||||||
|
texts: Union[List[str], np.ndarray],
|
||||||
|
*args,
|
||||||
|
**kwargs,
|
||||||
|
) -> List[List[float]]:
|
||||||
|
return self._watsonx_client.embed_documents(
|
||||||
|
texts=list(texts),
|
||||||
|
*args,
|
||||||
|
**kwargs,
|
||||||
|
)
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def _watsonx_client(self):
|
||||||
|
ibm_watsonx_ai = attempt_import_or_raise("ibm_watsonx_ai")
|
||||||
|
ibm_watsonx_ai_foundation_models = attempt_import_or_raise(
|
||||||
|
"ibm_watsonx_ai.foundation_models"
|
||||||
|
)
|
||||||
|
|
||||||
|
kwargs = {"model_id": self.name}
|
||||||
|
if self.params:
|
||||||
|
kwargs["params"] = self.params
|
||||||
|
if self.project_id:
|
||||||
|
kwargs["project_id"] = self.project_id
|
||||||
|
elif "WATSONX_PROJECT_ID" in os.environ:
|
||||||
|
kwargs["project_id"] = os.environ["WATSONX_PROJECT_ID"]
|
||||||
|
else:
|
||||||
|
raise ValueError("WATSONX_PROJECT_ID must be set or passed")
|
||||||
|
|
||||||
|
creds_kwargs = {}
|
||||||
|
if self.api_key:
|
||||||
|
creds_kwargs["api_key"] = self.api_key
|
||||||
|
elif "WATSONX_API_KEY" in os.environ:
|
||||||
|
creds_kwargs["api_key"] = os.environ["WATSONX_API_KEY"]
|
||||||
|
else:
|
||||||
|
raise ValueError("WATSONX_API_KEY must be set or passed")
|
||||||
|
if self.url:
|
||||||
|
creds_kwargs["url"] = self.url
|
||||||
|
else:
|
||||||
|
creds_kwargs["url"] = DEFAULT_WATSONX_URL
|
||||||
|
kwargs["credentials"] = ibm_watsonx_ai.Credentials(**creds_kwargs)
|
||||||
|
|
||||||
|
return ibm_watsonx_ai_foundation_models.Embeddings(**kwargs)
|
||||||
@@ -8,7 +8,7 @@ from ._lancedb import (
|
|||||||
)
|
)
|
||||||
|
|
||||||
|
|
||||||
class BTree(object):
|
class BTree:
|
||||||
"""Describes a btree index configuration
|
"""Describes a btree index configuration
|
||||||
|
|
||||||
A btree index is an index on scalar columns. The index stores a copy of the
|
A btree index is an index on scalar columns. The index stores a copy of the
|
||||||
@@ -22,7 +22,8 @@ class BTree(object):
|
|||||||
sizeof(Scalar) * 4096 bytes to find the correct row ids.
|
sizeof(Scalar) * 4096 bytes to find the correct row ids.
|
||||||
|
|
||||||
This index is good for scalar columns with mostly distinct values and does best
|
This index is good for scalar columns with mostly distinct values and does best
|
||||||
when the query is highly selective.
|
when the query is highly selective. It works with numeric, temporal, and string
|
||||||
|
columns.
|
||||||
|
|
||||||
The btree index does not currently have any parameters though parameters such as
|
The btree index does not currently have any parameters though parameters such as
|
||||||
the block size may be added in the future.
|
the block size may be added in the future.
|
||||||
@@ -32,7 +33,44 @@ class BTree(object):
|
|||||||
self._inner = LanceDbIndex.btree()
|
self._inner = LanceDbIndex.btree()
|
||||||
|
|
||||||
|
|
||||||
class IvfPq(object):
|
class Bitmap:
|
||||||
|
"""Describe a Bitmap index configuration.
|
||||||
|
|
||||||
|
A `Bitmap` index stores a bitmap for each distinct value in the column for
|
||||||
|
every row.
|
||||||
|
|
||||||
|
This index works best for low-cardinality numeric or string columns,
|
||||||
|
where the number of unique values is small (i.e., less than a few thousands).
|
||||||
|
`Bitmap` index can accelerate the following filters:
|
||||||
|
|
||||||
|
- `<`, `<=`, `=`, `>`, `>=`
|
||||||
|
- `IN (value1, value2, ...)`
|
||||||
|
- `between (value1, value2)`
|
||||||
|
- `is null`
|
||||||
|
|
||||||
|
For example, a bitmap index with a table with 1Bi rows, and 128 distinct values,
|
||||||
|
requires 128 / 8 * 1Bi bytes on disk.
|
||||||
|
"""
|
||||||
|
|
||||||
|
def __init__(self):
|
||||||
|
self._inner = LanceDbIndex.bitmap()
|
||||||
|
|
||||||
|
|
||||||
|
class LabelList:
|
||||||
|
"""Describe a LabelList index configuration.
|
||||||
|
|
||||||
|
`LabelList` is a scalar index that can be used on `List<T>` columns to
|
||||||
|
support queries with `array_contains_all` and `array_contains_any`
|
||||||
|
using an underlying bitmap index.
|
||||||
|
|
||||||
|
For example, it works with `tags`, `categories`, `keywords`, etc.
|
||||||
|
"""
|
||||||
|
|
||||||
|
def __init__(self):
|
||||||
|
self._inner = LanceDbIndex.label_list()
|
||||||
|
|
||||||
|
|
||||||
|
class IvfPq:
|
||||||
"""Describes an IVF PQ Index
|
"""Describes an IVF PQ Index
|
||||||
|
|
||||||
This index stores a compressed (quantized) copy of every vector. These vectors
|
This index stores a compressed (quantized) copy of every vector. These vectors
|
||||||
|
|||||||
@@ -99,6 +99,9 @@ class Query(pydantic.BaseModel):
|
|||||||
# if True then apply the filter before vector search
|
# if True then apply the filter before vector search
|
||||||
prefilter: bool = False
|
prefilter: bool = False
|
||||||
|
|
||||||
|
# full text search query
|
||||||
|
full_text_query: Optional[Union[str, dict]] = None
|
||||||
|
|
||||||
# top k results to return
|
# top k results to return
|
||||||
k: int
|
k: int
|
||||||
|
|
||||||
@@ -131,6 +134,7 @@ class LanceQueryBuilder(ABC):
|
|||||||
query_type: str,
|
query_type: str,
|
||||||
vector_column_name: str,
|
vector_column_name: str,
|
||||||
ordering_field_name: str = None,
|
ordering_field_name: str = None,
|
||||||
|
fts_columns: Union[str, List[str]] = None,
|
||||||
) -> LanceQueryBuilder:
|
) -> LanceQueryBuilder:
|
||||||
"""
|
"""
|
||||||
Create a query builder based on the given query and query type.
|
Create a query builder based on the given query and query type.
|
||||||
@@ -226,6 +230,7 @@ class LanceQueryBuilder(ABC):
|
|||||||
self._limit = 10
|
self._limit = 10
|
||||||
self._columns = None
|
self._columns = None
|
||||||
self._where = None
|
self._where = None
|
||||||
|
self._prefilter = False
|
||||||
self._with_row_id = False
|
self._with_row_id = False
|
||||||
|
|
||||||
@deprecation.deprecated(
|
@deprecation.deprecated(
|
||||||
@@ -428,9 +433,9 @@ class LanceQueryBuilder(ABC):
|
|||||||
>>> query = [100, 100]
|
>>> query = [100, 100]
|
||||||
>>> plan = table.search(query).explain_plan(True)
|
>>> plan = table.search(query).explain_plan(True)
|
||||||
>>> print(plan) # doctest: +ELLIPSIS, +NORMALIZE_WHITESPACE
|
>>> print(plan) # doctest: +ELLIPSIS, +NORMALIZE_WHITESPACE
|
||||||
Projection: fields=[vector, _distance]
|
ProjectionExec: expr=[vector@0 as vector, _distance@2 as _distance]
|
||||||
FilterExec: _distance@2 IS NOT NULL
|
FilterExec: _distance@2 IS NOT NULL
|
||||||
SortExec: TopK(fetch=10), expr=[_distance@2 ASC NULLS LAST]
|
SortExec: TopK(fetch=10), expr=[_distance@2 ASC NULLS LAST], preserve_partitioning=[false]
|
||||||
KNNVectorDistance: metric=l2
|
KNNVectorDistance: metric=l2
|
||||||
LanceScan: uri=..., projection=[vector], row_id=true, row_addr=false, ordered=false
|
LanceScan: uri=..., projection=[vector], row_id=true, row_addr=false, ordered=false
|
||||||
|
|
||||||
@@ -664,12 +669,19 @@ class LanceVectorQueryBuilder(LanceQueryBuilder):
|
|||||||
class LanceFtsQueryBuilder(LanceQueryBuilder):
|
class LanceFtsQueryBuilder(LanceQueryBuilder):
|
||||||
"""A builder for full text search for LanceDB."""
|
"""A builder for full text search for LanceDB."""
|
||||||
|
|
||||||
def __init__(self, table: "Table", query: str, ordering_field_name: str = None):
|
def __init__(
|
||||||
|
self,
|
||||||
|
table: "Table",
|
||||||
|
query: str,
|
||||||
|
ordering_field_name: str = None,
|
||||||
|
fts_columns: Union[str, List[str]] = None,
|
||||||
|
):
|
||||||
super().__init__(table)
|
super().__init__(table)
|
||||||
self._query = query
|
self._query = query
|
||||||
self._phrase_query = False
|
self._phrase_query = False
|
||||||
self.ordering_field_name = ordering_field_name
|
self.ordering_field_name = ordering_field_name
|
||||||
self._reranker = None
|
self._reranker = None
|
||||||
|
self._fts_columns = fts_columns
|
||||||
|
|
||||||
def phrase_query(self, phrase_query: bool = True) -> LanceFtsQueryBuilder:
|
def phrase_query(self, phrase_query: bool = True) -> LanceFtsQueryBuilder:
|
||||||
"""Set whether to use phrase query.
|
"""Set whether to use phrase query.
|
||||||
@@ -689,6 +701,35 @@ class LanceFtsQueryBuilder(LanceQueryBuilder):
|
|||||||
return self
|
return self
|
||||||
|
|
||||||
def to_arrow(self) -> pa.Table:
|
def to_arrow(self) -> pa.Table:
|
||||||
|
tantivy_index_path = self._table._get_fts_index_path()
|
||||||
|
if Path(tantivy_index_path).exists():
|
||||||
|
return self.tantivy_to_arrow()
|
||||||
|
|
||||||
|
query = self._query
|
||||||
|
if self._phrase_query:
|
||||||
|
raise NotImplementedError(
|
||||||
|
"Phrase query is not yet supported in Lance FTS. "
|
||||||
|
"Use tantivy-based index instead for now."
|
||||||
|
)
|
||||||
|
if self._reranker:
|
||||||
|
raise NotImplementedError(
|
||||||
|
"Reranking is not yet supported in Lance FTS. "
|
||||||
|
"Use tantivy-based index instead for now."
|
||||||
|
)
|
||||||
|
ds = self._table.to_lance()
|
||||||
|
return ds.to_table(
|
||||||
|
columns=self._columns,
|
||||||
|
filter=self._where,
|
||||||
|
limit=self._limit,
|
||||||
|
prefilter=self._prefilter,
|
||||||
|
with_row_id=self._with_row_id,
|
||||||
|
full_text_query={
|
||||||
|
"query": query,
|
||||||
|
"columns": self._fts_columns,
|
||||||
|
},
|
||||||
|
)
|
||||||
|
|
||||||
|
def tantivy_to_arrow(self) -> pa.Table:
|
||||||
try:
|
try:
|
||||||
import tantivy
|
import tantivy
|
||||||
except ImportError:
|
except ImportError:
|
||||||
@@ -726,11 +767,11 @@ class LanceFtsQueryBuilder(LanceQueryBuilder):
|
|||||||
index, query, self._limit, ordering_field=self.ordering_field_name
|
index, query, self._limit, ordering_field=self.ordering_field_name
|
||||||
)
|
)
|
||||||
if len(row_ids) == 0:
|
if len(row_ids) == 0:
|
||||||
empty_schema = pa.schema([pa.field("score", pa.float32())])
|
empty_schema = pa.schema([pa.field("_score", pa.float32())])
|
||||||
return pa.Table.from_pylist([], schema=empty_schema)
|
return pa.Table.from_pylist([], schema=empty_schema)
|
||||||
scores = pa.array(scores)
|
scores = pa.array(scores)
|
||||||
output_tbl = self._table.to_lance().take(row_ids, columns=self._columns)
|
output_tbl = self._table.to_lance().take(row_ids, columns=self._columns)
|
||||||
output_tbl = output_tbl.append_column("score", scores)
|
output_tbl = output_tbl.append_column("_score", scores)
|
||||||
# this needs to match vector search results which are uint64
|
# this needs to match vector search results which are uint64
|
||||||
row_ids = pa.array(row_ids, type=pa.uint64())
|
row_ids = pa.array(row_ids, type=pa.uint64())
|
||||||
|
|
||||||
@@ -784,8 +825,7 @@ class LanceFtsQueryBuilder(LanceQueryBuilder):
|
|||||||
LanceFtsQueryBuilder
|
LanceFtsQueryBuilder
|
||||||
The LanceQueryBuilder object.
|
The LanceQueryBuilder object.
|
||||||
"""
|
"""
|
||||||
self._reranker = reranker
|
raise NotImplementedError("Reranking is not yet supported for FTS queries.")
|
||||||
return self
|
|
||||||
|
|
||||||
|
|
||||||
class LanceEmptyQueryBuilder(LanceQueryBuilder):
|
class LanceEmptyQueryBuilder(LanceQueryBuilder):
|
||||||
@@ -856,13 +896,13 @@ class LanceHybridQueryBuilder(LanceQueryBuilder):
|
|||||||
# convert to ranks first if needed
|
# convert to ranks first if needed
|
||||||
if self._norm == "rank":
|
if self._norm == "rank":
|
||||||
vector_results = self._rank(vector_results, "_distance")
|
vector_results = self._rank(vector_results, "_distance")
|
||||||
fts_results = self._rank(fts_results, "score")
|
fts_results = self._rank(fts_results, "_score")
|
||||||
# normalize the scores to be between 0 and 1, 0 being most relevant
|
# normalize the scores to be between 0 and 1, 0 being most relevant
|
||||||
vector_results = self._normalize_scores(vector_results, "_distance")
|
vector_results = self._normalize_scores(vector_results, "_distance")
|
||||||
|
|
||||||
# In fts higher scores represent relevance. Not inverting them here as
|
# In fts higher scores represent relevance. Not inverting them here as
|
||||||
# rerankers might need to preserve this score to support `return_score="all"`
|
# rerankers might need to preserve this score to support `return_score="all"`
|
||||||
fts_results = self._normalize_scores(fts_results, "score")
|
fts_results = self._normalize_scores(fts_results, "_score")
|
||||||
|
|
||||||
results = self._reranker.rerank_hybrid(
|
results = self._reranker.rerank_hybrid(
|
||||||
self._fts_query._query, vector_results, fts_results
|
self._fts_query._query, vector_results, fts_results
|
||||||
@@ -1177,6 +1217,16 @@ class AsyncQueryBase(object):
|
|||||||
await batch_iter.read_all(), schema=batch_iter.schema
|
await batch_iter.read_all(), schema=batch_iter.schema
|
||||||
)
|
)
|
||||||
|
|
||||||
|
async def to_list(self) -> List[dict]:
|
||||||
|
"""
|
||||||
|
Execute the query and return the results as a list of dictionaries.
|
||||||
|
|
||||||
|
Each list entry is a dictionary with the selected column names as keys,
|
||||||
|
or all table columns if `select` is not called. The vector and the "_distance"
|
||||||
|
fields are returned whether or not they're explicitly selected.
|
||||||
|
"""
|
||||||
|
return (await self.to_arrow()).to_pylist()
|
||||||
|
|
||||||
async def to_pandas(self) -> "pd.DataFrame":
|
async def to_pandas(self) -> "pd.DataFrame":
|
||||||
"""
|
"""
|
||||||
Execute the query and collect the results into a pandas DataFrame.
|
Execute the query and collect the results into a pandas DataFrame.
|
||||||
@@ -1214,9 +1264,9 @@ class AsyncQueryBase(object):
|
|||||||
... plan = await table.query().nearest_to([1, 2]).explain_plan(True)
|
... plan = await table.query().nearest_to([1, 2]).explain_plan(True)
|
||||||
... print(plan)
|
... print(plan)
|
||||||
>>> asyncio.run(doctest_example()) # doctest: +ELLIPSIS, +NORMALIZE_WHITESPACE
|
>>> asyncio.run(doctest_example()) # doctest: +ELLIPSIS, +NORMALIZE_WHITESPACE
|
||||||
Projection: fields=[vector, _distance]
|
ProjectionExec: expr=[vector@0 as vector, _distance@2 as _distance]
|
||||||
FilterExec: _distance@2 IS NOT NULL
|
FilterExec: _distance@2 IS NOT NULL
|
||||||
SortExec: TopK(fetch=10), expr=[_distance@2 ASC NULLS LAST]
|
SortExec: TopK(fetch=10), expr=[_distance@2 ASC NULLS LAST], preserve_partitioning=[false]
|
||||||
KNNVectorDistance: metric=l2
|
KNNVectorDistance: metric=l2
|
||||||
LanceScan: uri=..., projection=[vector], row_id=true, row_addr=false, ordered=false
|
LanceScan: uri=..., projection=[vector], row_id=true, row_addr=false, ordered=false
|
||||||
|
|
||||||
|
|||||||
@@ -245,7 +245,7 @@ class RemoteDBConnection(DBConnection):
|
|||||||
schema = schema.to_arrow_schema()
|
schema = schema.to_arrow_schema()
|
||||||
|
|
||||||
if data is not None:
|
if data is not None:
|
||||||
data = _sanitize_data(
|
data, schema = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
schema,
|
schema,
|
||||||
metadata=None,
|
metadata=None,
|
||||||
|
|||||||
@@ -22,8 +22,9 @@ from lance import json_to_schema
|
|||||||
|
|
||||||
from lancedb.common import DATA, VEC, VECTOR_COLUMN_NAME
|
from lancedb.common import DATA, VEC, VECTOR_COLUMN_NAME
|
||||||
from lancedb.merge import LanceMergeInsertBuilder
|
from lancedb.merge import LanceMergeInsertBuilder
|
||||||
|
from lancedb.embeddings import EmbeddingFunctionRegistry
|
||||||
|
|
||||||
from ..query import LanceVectorQueryBuilder
|
from ..query import LanceVectorQueryBuilder, LanceQueryBuilder
|
||||||
from ..table import Query, Table, _sanitize_data
|
from ..table import Query, Table, _sanitize_data
|
||||||
from ..util import inf_vector_column_query, value_to_sql
|
from ..util import inf_vector_column_query, value_to_sql
|
||||||
from .arrow import to_ipc_binary
|
from .arrow import to_ipc_binary
|
||||||
@@ -58,6 +59,21 @@ class RemoteTable(Table):
|
|||||||
resp = self._conn._client.post(f"/v1/table/{self._name}/describe/")
|
resp = self._conn._client.post(f"/v1/table/{self._name}/describe/")
|
||||||
return resp["version"]
|
return resp["version"]
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def embedding_functions(self) -> dict:
|
||||||
|
"""
|
||||||
|
Get the embedding functions for the table
|
||||||
|
|
||||||
|
Returns
|
||||||
|
-------
|
||||||
|
funcs: dict
|
||||||
|
A mapping of the vector column to the embedding function
|
||||||
|
or empty dict if not configured.
|
||||||
|
"""
|
||||||
|
return EmbeddingFunctionRegistry.get_instance().parse_functions(
|
||||||
|
self.schema.metadata
|
||||||
|
)
|
||||||
|
|
||||||
def to_arrow(self) -> pa.Table:
|
def to_arrow(self) -> pa.Table:
|
||||||
"""to_arrow() is not yet supported on LanceDB cloud."""
|
"""to_arrow() is not yet supported on LanceDB cloud."""
|
||||||
raise NotImplementedError("to_arrow() is not yet supported on LanceDB cloud.")
|
raise NotImplementedError("to_arrow() is not yet supported on LanceDB cloud.")
|
||||||
@@ -210,10 +226,10 @@ class RemoteTable(Table):
|
|||||||
The value to use when filling vectors. Only used if on_bad_vectors="fill".
|
The value to use when filling vectors. Only used if on_bad_vectors="fill".
|
||||||
|
|
||||||
"""
|
"""
|
||||||
data = _sanitize_data(
|
data, _ = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
self.schema,
|
self.schema,
|
||||||
metadata=None,
|
metadata=self.schema.metadata,
|
||||||
on_bad_vectors=on_bad_vectors,
|
on_bad_vectors=on_bad_vectors,
|
||||||
fill_value=fill_value,
|
fill_value=fill_value,
|
||||||
)
|
)
|
||||||
@@ -293,6 +309,7 @@ class RemoteTable(Table):
|
|||||||
"""
|
"""
|
||||||
if vector_column_name is None:
|
if vector_column_name is None:
|
||||||
vector_column_name = inf_vector_column_query(self.schema)
|
vector_column_name = inf_vector_column_query(self.schema)
|
||||||
|
query = LanceQueryBuilder._query_to_vector(self, query, vector_column_name)
|
||||||
return LanceVectorQueryBuilder(self, query, vector_column_name)
|
return LanceVectorQueryBuilder(self, query, vector_column_name)
|
||||||
|
|
||||||
def _execute_query(
|
def _execute_query(
|
||||||
@@ -336,7 +353,7 @@ class RemoteTable(Table):
|
|||||||
|
|
||||||
See [`Table.merge_insert`][lancedb.table.Table.merge_insert] for more details.
|
See [`Table.merge_insert`][lancedb.table.Table.merge_insert] for more details.
|
||||||
"""
|
"""
|
||||||
super().merge_insert(on)
|
return super().merge_insert(on)
|
||||||
|
|
||||||
def _do_merge(
|
def _do_merge(
|
||||||
self,
|
self,
|
||||||
@@ -345,7 +362,7 @@ class RemoteTable(Table):
|
|||||||
on_bad_vectors: str,
|
on_bad_vectors: str,
|
||||||
fill_value: float,
|
fill_value: float,
|
||||||
):
|
):
|
||||||
data = _sanitize_data(
|
data, _ = _sanitize_data(
|
||||||
new_data,
|
new_data,
|
||||||
self.schema,
|
self.schema,
|
||||||
metadata=None,
|
metadata=None,
|
||||||
|
|||||||
@@ -5,6 +5,7 @@ from .cross_encoder import CrossEncoderReranker
|
|||||||
from .linear_combination import LinearCombinationReranker
|
from .linear_combination import LinearCombinationReranker
|
||||||
from .openai import OpenaiReranker
|
from .openai import OpenaiReranker
|
||||||
from .jinaai import JinaReranker
|
from .jinaai import JinaReranker
|
||||||
|
from .rrf import RRFReranker
|
||||||
|
|
||||||
__all__ = [
|
__all__ = [
|
||||||
"Reranker",
|
"Reranker",
|
||||||
@@ -14,4 +15,5 @@ __all__ = [
|
|||||||
"OpenaiReranker",
|
"OpenaiReranker",
|
||||||
"ColbertReranker",
|
"ColbertReranker",
|
||||||
"JinaReranker",
|
"JinaReranker",
|
||||||
|
"RRFReranker",
|
||||||
]
|
]
|
||||||
|
|||||||
@@ -1,9 +1,13 @@
|
|||||||
from abc import ABC, abstractmethod
|
from abc import ABC, abstractmethod
|
||||||
from packaging.version import Version
|
from packaging.version import Version
|
||||||
|
from typing import Union, List, TYPE_CHECKING
|
||||||
|
|
||||||
import numpy as np
|
import numpy as np
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
|
|
||||||
|
if TYPE_CHECKING:
|
||||||
|
from ..table import LanceVectorQueryBuilder
|
||||||
|
|
||||||
ARROW_VERSION = Version(pa.__version__)
|
ARROW_VERSION = Version(pa.__version__)
|
||||||
|
|
||||||
|
|
||||||
@@ -130,12 +134,94 @@ class Reranker(ABC):
|
|||||||
combined = pa.concat_tables(
|
combined = pa.concat_tables(
|
||||||
[vector_results, fts_results], **self._concat_tables_args
|
[vector_results, fts_results], **self._concat_tables_args
|
||||||
)
|
)
|
||||||
row_id = combined.column("_rowid")
|
|
||||||
|
|
||||||
# deduplicate
|
# deduplicate
|
||||||
mask = np.full((combined.shape[0]), False)
|
combined = self._deduplicate(combined)
|
||||||
_, mask_indices = np.unique(np.array(row_id), return_index=True)
|
|
||||||
mask[mask_indices] = True
|
|
||||||
combined = combined.filter(mask=mask)
|
|
||||||
|
|
||||||
return combined
|
return combined
|
||||||
|
|
||||||
|
def rerank_multivector(
|
||||||
|
self,
|
||||||
|
vector_results: Union[List[pa.Table], List["LanceVectorQueryBuilder"]],
|
||||||
|
query: Union[str, None], # Some rerankers might not need the query
|
||||||
|
deduplicate: bool = False,
|
||||||
|
):
|
||||||
|
"""
|
||||||
|
This is a rerank function that receives the results from multiple
|
||||||
|
vector searches. For example, this can be used to combine the
|
||||||
|
results of two vector searches with different embeddings.
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
vector_results : List[pa.Table] or List[LanceVectorQueryBuilder]
|
||||||
|
The results from the vector search. Either accepts the query builder
|
||||||
|
if the results haven't been executed yet or the results in arrow format.
|
||||||
|
query : str or None,
|
||||||
|
The input query. Some rerankers might not need the query to rerank.
|
||||||
|
In that case, it can be set to None explicitly. This is inteded to
|
||||||
|
be handled by the reranker implementations.
|
||||||
|
deduplicate : bool, optional
|
||||||
|
Whether to deduplicate the results based on the `_rowid` column,
|
||||||
|
by default False. Requires `_rowid` to be present in the results.
|
||||||
|
|
||||||
|
Returns
|
||||||
|
-------
|
||||||
|
pa.Table
|
||||||
|
The reranked results
|
||||||
|
"""
|
||||||
|
vector_results = (
|
||||||
|
[vector_results] if not isinstance(vector_results, list) else vector_results
|
||||||
|
)
|
||||||
|
|
||||||
|
# Make sure all elements are of the same type
|
||||||
|
if not all(isinstance(v, type(vector_results[0])) for v in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"All elements in vector_results should be of the same type"
|
||||||
|
)
|
||||||
|
|
||||||
|
# avoids circular import
|
||||||
|
if type(vector_results[0]).__name__ == "LanceVectorQueryBuilder":
|
||||||
|
vector_results = [result.to_arrow() for result in vector_results]
|
||||||
|
elif not isinstance(vector_results[0], pa.Table):
|
||||||
|
raise ValueError(
|
||||||
|
"vector_results should be a list of pa.Table or LanceVectorQueryBuilder"
|
||||||
|
)
|
||||||
|
|
||||||
|
combined = pa.concat_tables(vector_results, **self._concat_tables_args)
|
||||||
|
|
||||||
|
reranked = self.rerank_vector(query, combined)
|
||||||
|
|
||||||
|
# TODO: Allow custom deduplicators here.
|
||||||
|
# currently, this'll just keep the first instance.
|
||||||
|
if deduplicate:
|
||||||
|
if "_rowid" not in combined.column_names:
|
||||||
|
raise ValueError(
|
||||||
|
"'_rowid' is required for deduplication. \
|
||||||
|
add _rowid to search results like this: \
|
||||||
|
`search().with_row_id(True)`"
|
||||||
|
)
|
||||||
|
reranked = self._deduplicate(reranked)
|
||||||
|
|
||||||
|
return reranked
|
||||||
|
|
||||||
|
def _deduplicate(self, table: pa.Table):
|
||||||
|
"""
|
||||||
|
Deduplicate the table based on the `_rowid` column.
|
||||||
|
"""
|
||||||
|
row_id = table.column("_rowid")
|
||||||
|
|
||||||
|
# deduplicate
|
||||||
|
mask = np.full((table.shape[0]), False)
|
||||||
|
_, mask_indices = np.unique(np.array(row_id), return_index=True)
|
||||||
|
mask[mask_indices] = True
|
||||||
|
deduped_table = table.filter(mask=mask)
|
||||||
|
|
||||||
|
return deduped_table
|
||||||
|
|
||||||
|
def _keep_relevance_score(self, combined_results: pa.Table):
|
||||||
|
if self.score == "relevance":
|
||||||
|
if "_score" in combined_results.column_names:
|
||||||
|
combined_results = combined_results.drop_columns(["_score"])
|
||||||
|
if "_distance" in combined_results.column_names:
|
||||||
|
combined_results = combined_results.drop_columns(["_distance"])
|
||||||
|
return combined_results
|
||||||
|
|||||||
@@ -88,7 +88,7 @@ class CohereReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for cohere reranker"
|
"return_score='all' not implemented for cohere reranker"
|
||||||
@@ -113,6 +113,6 @@ class CohereReranker(Reranker):
|
|||||||
):
|
):
|
||||||
result_set = self._rerank(fts_results, query)
|
result_set = self._rerank(fts_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
result_set = result_set.drop_columns(["score"])
|
result_set = result_set.drop_columns(["_score"])
|
||||||
|
|
||||||
return result_set
|
return result_set
|
||||||
|
|||||||
@@ -73,7 +73,7 @@ class ColbertReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"OpenAI Reranker does not support score='all' yet"
|
"OpenAI Reranker does not support score='all' yet"
|
||||||
@@ -105,7 +105,7 @@ class ColbertReranker(Reranker):
|
|||||||
):
|
):
|
||||||
result_set = self._rerank(fts_results, query)
|
result_set = self._rerank(fts_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
result_set = result_set.drop_columns(["score"])
|
result_set = result_set.drop_columns(["_score"])
|
||||||
|
|
||||||
result_set = result_set.sort_by([("_relevance_score", "descending")])
|
result_set = result_set.sort_by([("_relevance_score", "descending")])
|
||||||
|
|
||||||
|
|||||||
@@ -66,7 +66,7 @@ class CrossEncoderReranker(Reranker):
|
|||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
# sort the results by _score
|
# sort the results by _score
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for CrossEncoderReranker"
|
"return_score='all' not implemented for CrossEncoderReranker"
|
||||||
@@ -96,7 +96,7 @@ class CrossEncoderReranker(Reranker):
|
|||||||
):
|
):
|
||||||
fts_results = self._rerank(fts_results, query)
|
fts_results = self._rerank(fts_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
fts_results = fts_results.drop_columns(["score"])
|
fts_results = fts_results.drop_columns(["_score"])
|
||||||
|
|
||||||
fts_results = fts_results.sort_by([("_relevance_score", "descending")])
|
fts_results = fts_results.sort_by([("_relevance_score", "descending")])
|
||||||
return fts_results
|
return fts_results
|
||||||
|
|||||||
@@ -92,7 +92,7 @@ class JinaReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for JinaReranker"
|
"return_score='all' not implemented for JinaReranker"
|
||||||
@@ -117,6 +117,6 @@ class JinaReranker(Reranker):
|
|||||||
):
|
):
|
||||||
result_set = self._rerank(fts_results, query)
|
result_set = self._rerank(fts_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
result_set = result_set.drop_columns(["score"])
|
result_set = result_set.drop_columns(["_score"])
|
||||||
|
|
||||||
return result_set
|
return result_set
|
||||||
|
|||||||
@@ -69,12 +69,12 @@ class LinearCombinationReranker(Reranker):
|
|||||||
vi = vector_list[i]
|
vi = vector_list[i]
|
||||||
fj = fts_list[j]
|
fj = fts_list[j]
|
||||||
# invert the fts score from relevance to distance
|
# invert the fts score from relevance to distance
|
||||||
inverted_fts_score = self._invert_score(fj["score"])
|
inverted_fts_score = self._invert_score(fj["_score"])
|
||||||
if vi["_rowid"] == fj["_rowid"]:
|
if vi["_rowid"] == fj["_rowid"]:
|
||||||
vi["_relevance_score"] = self._combine_score(
|
vi["_relevance_score"] = self._combine_score(
|
||||||
vi["_distance"], inverted_fts_score
|
vi["_distance"], inverted_fts_score
|
||||||
)
|
)
|
||||||
vi["score"] = fj["score"] # keep the original score
|
vi["_score"] = fj["_score"] # keep the original score
|
||||||
combined_list.append(vi)
|
combined_list.append(vi)
|
||||||
i += 1
|
i += 1
|
||||||
j += 1
|
j += 1
|
||||||
@@ -103,7 +103,7 @@ class LinearCombinationReranker(Reranker):
|
|||||||
[("_relevance_score", "descending")]
|
[("_relevance_score", "descending")]
|
||||||
)
|
)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
tbl = tbl.drop_columns(["score", "_distance"])
|
tbl = self._keep_relevance_score(tbl)
|
||||||
return tbl
|
return tbl
|
||||||
|
|
||||||
def _combine_score(self, score1, score2):
|
def _combine_score(self, score1, score2):
|
||||||
|
|||||||
@@ -84,7 +84,7 @@ class OpenaiReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"OpenAI Reranker does not support score='all' yet"
|
"OpenAI Reranker does not support score='all' yet"
|
||||||
@@ -108,7 +108,7 @@ class OpenaiReranker(Reranker):
|
|||||||
def rerank_fts(self, query: str, fts_results: pa.Table):
|
def rerank_fts(self, query: str, fts_results: pa.Table):
|
||||||
fts_results = self._rerank(fts_results, query)
|
fts_results = self._rerank(fts_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
fts_results = fts_results.drop_columns(["score"])
|
fts_results = fts_results.drop_columns(["_score"])
|
||||||
|
|
||||||
fts_results = fts_results.sort_by([("_relevance_score", "descending")])
|
fts_results = fts_results.sort_by([("_relevance_score", "descending")])
|
||||||
|
|
||||||
|
|||||||
104
python/python/lancedb/rerankers/rrf.py
Normal file
104
python/python/lancedb/rerankers/rrf.py
Normal file
@@ -0,0 +1,104 @@
|
|||||||
|
from typing import Union, List, TYPE_CHECKING
|
||||||
|
import pyarrow as pa
|
||||||
|
|
||||||
|
from collections import defaultdict
|
||||||
|
from .base import Reranker
|
||||||
|
|
||||||
|
if TYPE_CHECKING:
|
||||||
|
from ..table import LanceVectorQueryBuilder
|
||||||
|
|
||||||
|
|
||||||
|
class RRFReranker(Reranker):
|
||||||
|
"""
|
||||||
|
Reranks the results using Reciprocal Rank Fusion(RRF) algorithm based
|
||||||
|
on the scores of vector and FTS search.
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
K : int, default 60
|
||||||
|
A constant used in the RRF formula (default is 60). Experiments
|
||||||
|
indicate that k = 60 was near-optimal, but that the choice is
|
||||||
|
not critical. See paper:
|
||||||
|
https://plg.uwaterloo.ca/~gvcormac/cormacksigir09-rrf.pdf
|
||||||
|
return_score : str, default "relevance"
|
||||||
|
opntions are "relevance" or "all"
|
||||||
|
The type of score to return. If "relevance", will return only the relevance
|
||||||
|
score. If "all", will return all scores from the vector and FTS search along
|
||||||
|
with the relevance score.
|
||||||
|
"""
|
||||||
|
|
||||||
|
def __init__(self, K: int = 60, return_score="relevance"):
|
||||||
|
if K <= 0:
|
||||||
|
raise ValueError("K must be greater than 0")
|
||||||
|
super().__init__(return_score)
|
||||||
|
self.K = K
|
||||||
|
|
||||||
|
def rerank_hybrid(
|
||||||
|
self,
|
||||||
|
query: str, # noqa: F821
|
||||||
|
vector_results: pa.Table,
|
||||||
|
fts_results: pa.Table,
|
||||||
|
):
|
||||||
|
vector_ids = vector_results["_rowid"].to_pylist() if vector_results else []
|
||||||
|
fts_ids = fts_results["_rowid"].to_pylist() if fts_results else []
|
||||||
|
rrf_score_map = defaultdict(float)
|
||||||
|
|
||||||
|
# Calculate RRF score of each result
|
||||||
|
for ids in [vector_ids, fts_ids]:
|
||||||
|
for i, result_id in enumerate(ids, 1):
|
||||||
|
rrf_score_map[result_id] += 1 / (i + self.K)
|
||||||
|
|
||||||
|
# Sort the results based on RRF score
|
||||||
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
|
combined_row_ids = combined_results["_rowid"].to_pylist()
|
||||||
|
relevance_scores = [rrf_score_map[row_id] for row_id in combined_row_ids]
|
||||||
|
combined_results = combined_results.append_column(
|
||||||
|
"_relevance_score", pa.array(relevance_scores, type=pa.float32())
|
||||||
|
)
|
||||||
|
combined_results = combined_results.sort_by(
|
||||||
|
[("_relevance_score", "descending")]
|
||||||
|
)
|
||||||
|
|
||||||
|
if self.score == "relevance":
|
||||||
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
|
|
||||||
|
return combined_results
|
||||||
|
|
||||||
|
def rerank_multivector(
|
||||||
|
self,
|
||||||
|
vector_results: Union[List[pa.Table], List["LanceVectorQueryBuilder"]],
|
||||||
|
query: str = None,
|
||||||
|
deduplicate: bool = True, # noqa: F821 # TODO: automatically deduplicates
|
||||||
|
):
|
||||||
|
"""
|
||||||
|
Overridden method to rerank the results from multiple vector searches.
|
||||||
|
This leverages the RRF hybrid reranking algorithm to combine the
|
||||||
|
results from multiple vector searches as it doesn't support reranking
|
||||||
|
vector results individually.
|
||||||
|
"""
|
||||||
|
# Make sure all elements are of the same type
|
||||||
|
if not all(isinstance(v, type(vector_results[0])) for v in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"All elements in vector_results should be of the same type"
|
||||||
|
)
|
||||||
|
|
||||||
|
# avoid circular import
|
||||||
|
if type(vector_results[0]).__name__ == "LanceVectorQueryBuilder":
|
||||||
|
vector_results = [result.to_arrow() for result in vector_results]
|
||||||
|
elif not isinstance(vector_results[0], pa.Table):
|
||||||
|
raise ValueError(
|
||||||
|
"vector_results should be a list of pa.Table or LanceVectorQueryBuilder"
|
||||||
|
)
|
||||||
|
|
||||||
|
# _rowid is required for RRF reranking
|
||||||
|
if not all("_rowid" in result.column_names for result in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"'_rowid' is required for deduplication. \
|
||||||
|
add _rowid to search results like this: \
|
||||||
|
`search().with_row_id(True)`"
|
||||||
|
)
|
||||||
|
|
||||||
|
combined = pa.concat_tables(vector_results, **self._concat_tables_args)
|
||||||
|
empty_table = pa.Table.from_arrays([], names=[])
|
||||||
|
reranked = self.rerank_hybrid(query, combined, empty_table)
|
||||||
|
|
||||||
|
return reranked
|
||||||
@@ -1,15 +1,5 @@
|
|||||||
# Copyright 2023 LanceDB Developers
|
# SPDX-License-Identifier: Apache-2.0
|
||||||
#
|
# SPDX-FileCopyrightText: Copyright The LanceDB Authors
|
||||||
# Licensed under the Apache License, Version 2.0 (the "License");
|
|
||||||
# you may not use this file except in compliance with the License.
|
|
||||||
# You may obtain a copy of the License at
|
|
||||||
# http://www.apache.org/licenses/LICENSE-2.0
|
|
||||||
#
|
|
||||||
# Unless required by applicable law or agreed to in writing, software
|
|
||||||
# distributed under the License is distributed on an "AS IS" BASIS,
|
|
||||||
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
|
||||||
# See the License for the specific language governing permissions and
|
|
||||||
# limitations under the License.
|
|
||||||
|
|
||||||
from __future__ import annotations
|
from __future__ import annotations
|
||||||
|
|
||||||
@@ -59,10 +49,9 @@ from .util import (
|
|||||||
if TYPE_CHECKING:
|
if TYPE_CHECKING:
|
||||||
import PIL
|
import PIL
|
||||||
from lance.dataset import CleanupStats, ReaderLike
|
from lance.dataset import CleanupStats, ReaderLike
|
||||||
|
|
||||||
from ._lancedb import Table as LanceDBTable, OptimizeStats
|
from ._lancedb import Table as LanceDBTable, OptimizeStats
|
||||||
from .db import LanceDBConnection
|
from .db import LanceDBConnection
|
||||||
from .index import BTree, IndexConfig, IvfPq
|
from .index import BTree, IndexConfig, IvfPq, Bitmap, LabelList
|
||||||
|
|
||||||
|
|
||||||
pd = safe_import_pandas()
|
pd = safe_import_pandas()
|
||||||
@@ -103,6 +92,7 @@ def _sanitize_data(
|
|||||||
if isinstance(data, list):
|
if isinstance(data, list):
|
||||||
# convert to list of dict if data is a bunch of LanceModels
|
# convert to list of dict if data is a bunch of LanceModels
|
||||||
if isinstance(data[0], LanceModel):
|
if isinstance(data[0], LanceModel):
|
||||||
|
if schema is None:
|
||||||
schema = data[0].__class__.to_arrow_schema()
|
schema = data[0].__class__.to_arrow_schema()
|
||||||
data = [model_to_dict(d) for d in data]
|
data = [model_to_dict(d) for d in data]
|
||||||
data = pa.Table.from_pylist(data, schema=schema)
|
data = pa.Table.from_pylist(data, schema=schema)
|
||||||
@@ -133,7 +123,7 @@ def _sanitize_data(
|
|||||||
)
|
)
|
||||||
else:
|
else:
|
||||||
raise TypeError(f"Unsupported data type: {type(data)}")
|
raise TypeError(f"Unsupported data type: {type(data)}")
|
||||||
return data
|
return data, schema
|
||||||
|
|
||||||
|
|
||||||
def _schema_from_hf(data, schema):
|
def _schema_from_hf(data, schema):
|
||||||
@@ -205,7 +195,7 @@ def _to_record_batch_generator(
|
|||||||
# and do things like add the vector column etc
|
# and do things like add the vector column etc
|
||||||
if isinstance(batch, pa.RecordBatch):
|
if isinstance(batch, pa.RecordBatch):
|
||||||
batch = pa.Table.from_batches([batch])
|
batch = pa.Table.from_batches([batch])
|
||||||
batch = _sanitize_data(batch, schema, metadata, on_bad_vectors, fill_value)
|
batch, _ = _sanitize_data(batch, schema, metadata, on_bad_vectors, fill_value)
|
||||||
for b in batch.to_batches():
|
for b in batch.to_batches():
|
||||||
yield b
|
yield b
|
||||||
|
|
||||||
@@ -349,6 +339,7 @@ class Table(ABC):
|
|||||||
def create_scalar_index(
|
def create_scalar_index(
|
||||||
self,
|
self,
|
||||||
column: str,
|
column: str,
|
||||||
|
index_type: Literal["BTREE", "BITMAP", "LABEL_LIST"] = "BTREE",
|
||||||
*,
|
*,
|
||||||
replace: bool = True,
|
replace: bool = True,
|
||||||
):
|
):
|
||||||
@@ -510,6 +501,8 @@ class Table(ABC):
|
|||||||
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
vector_column_name: Optional[str] = None,
|
vector_column_name: Optional[str] = None,
|
||||||
query_type: str = "auto",
|
query_type: str = "auto",
|
||||||
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Union[str, List[str]] = None,
|
||||||
) -> LanceQueryBuilder:
|
) -> LanceQueryBuilder:
|
||||||
"""Create a search query to find the nearest neighbors
|
"""Create a search query to find the nearest neighbors
|
||||||
of the given query vector. We currently support [vector search][search]
|
of the given query vector. We currently support [vector search][search]
|
||||||
@@ -1187,9 +1180,15 @@ class LanceTable(Table):
|
|||||||
index_cache_size=index_cache_size,
|
index_cache_size=index_cache_size,
|
||||||
)
|
)
|
||||||
|
|
||||||
def create_scalar_index(self, column: str, *, replace: bool = True):
|
def create_scalar_index(
|
||||||
|
self,
|
||||||
|
column: str,
|
||||||
|
index_type: Literal["BTREE", "BITMAP", "LABEL_LIST"] = "BTREE",
|
||||||
|
*,
|
||||||
|
replace: bool = True,
|
||||||
|
):
|
||||||
self._dataset_mut.create_scalar_index(
|
self._dataset_mut.create_scalar_index(
|
||||||
column, index_type="BTREE", replace=replace
|
column, index_type=index_type, replace=replace
|
||||||
)
|
)
|
||||||
|
|
||||||
def create_fts_index(
|
def create_fts_index(
|
||||||
@@ -1200,6 +1199,7 @@ class LanceTable(Table):
|
|||||||
replace: bool = False,
|
replace: bool = False,
|
||||||
writer_heap_size: Optional[int] = 1024 * 1024 * 1024,
|
writer_heap_size: Optional[int] = 1024 * 1024 * 1024,
|
||||||
tokenizer_name: str = "default",
|
tokenizer_name: str = "default",
|
||||||
|
use_tantivy: bool = True,
|
||||||
):
|
):
|
||||||
"""Create a full-text search index on the table.
|
"""Create a full-text search index on the table.
|
||||||
|
|
||||||
@@ -1210,6 +1210,7 @@ class LanceTable(Table):
|
|||||||
----------
|
----------
|
||||||
field_names: str or list of str
|
field_names: str or list of str
|
||||||
The name(s) of the field to index.
|
The name(s) of the field to index.
|
||||||
|
can be only str if use_tantivy=True for now.
|
||||||
replace: bool, default False
|
replace: bool, default False
|
||||||
If True, replace the existing index if it exists. Note that this is
|
If True, replace the existing index if it exists. Note that this is
|
||||||
not yet an atomic operation; the index will be temporarily
|
not yet an atomic operation; the index will be temporarily
|
||||||
@@ -1217,12 +1218,31 @@ class LanceTable(Table):
|
|||||||
writer_heap_size: int, default 1GB
|
writer_heap_size: int, default 1GB
|
||||||
ordering_field_names:
|
ordering_field_names:
|
||||||
A list of unsigned type fields to index to optionally order
|
A list of unsigned type fields to index to optionally order
|
||||||
results on at search time
|
results on at search time.
|
||||||
|
only available with use_tantivy=True
|
||||||
tokenizer_name: str, default "default"
|
tokenizer_name: str, default "default"
|
||||||
The tokenizer to use for the index. Can be "raw", "default" or the 2 letter
|
The tokenizer to use for the index. Can be "raw", "default" or the 2 letter
|
||||||
language code followed by "_stem". So for english it would be "en_stem".
|
language code followed by "_stem". So for english it would be "en_stem".
|
||||||
For available languages see: https://docs.rs/tantivy/latest/tantivy/tokenizer/enum.Language.html
|
For available languages see: https://docs.rs/tantivy/latest/tantivy/tokenizer/enum.Language.html
|
||||||
|
only available with use_tantivy=True for now
|
||||||
|
use_tantivy: bool, default False
|
||||||
|
If True, use the legacy full-text search implementation based on tantivy.
|
||||||
|
If False, use the new full-text search implementation based on lance-index.
|
||||||
"""
|
"""
|
||||||
|
if not use_tantivy:
|
||||||
|
if not isinstance(field_names, str):
|
||||||
|
raise ValueError("field_names must be a string when use_tantivy=False")
|
||||||
|
# delete the existing legacy index if it exists
|
||||||
|
if replace:
|
||||||
|
fs, path = fs_from_uri(self._get_fts_index_path())
|
||||||
|
index_exists = fs.get_file_info(path).type != pa_fs.FileType.NotFound
|
||||||
|
if index_exists:
|
||||||
|
fs.delete_dir(path)
|
||||||
|
self._dataset_mut.create_scalar_index(
|
||||||
|
field_names, index_type="INVERTED", replace=replace
|
||||||
|
)
|
||||||
|
return
|
||||||
|
|
||||||
from .fts import create_index, populate_index
|
from .fts import create_index, populate_index
|
||||||
|
|
||||||
if isinstance(field_names, str):
|
if isinstance(field_names, str):
|
||||||
@@ -1295,7 +1315,7 @@ class LanceTable(Table):
|
|||||||
The number of vectors in the table.
|
The number of vectors in the table.
|
||||||
"""
|
"""
|
||||||
# TODO: manage table listing and metadata separately
|
# TODO: manage table listing and metadata separately
|
||||||
data = _sanitize_data(
|
data, _ = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
self.schema,
|
self.schema,
|
||||||
metadata=self.schema.metadata,
|
metadata=self.schema.metadata,
|
||||||
@@ -1391,6 +1411,7 @@ class LanceTable(Table):
|
|||||||
vector_column_name: Optional[str] = None,
|
vector_column_name: Optional[str] = None,
|
||||||
query_type: str = "auto",
|
query_type: str = "auto",
|
||||||
ordering_field_name: Optional[str] = None,
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Union[str, List[str]] = None,
|
||||||
) -> LanceQueryBuilder:
|
) -> LanceQueryBuilder:
|
||||||
"""Create a search query to find the nearest neighbors
|
"""Create a search query to find the nearest neighbors
|
||||||
of the given query vector. We currently support [vector search][search]
|
of the given query vector. We currently support [vector search][search]
|
||||||
@@ -1445,6 +1466,10 @@ class LanceTable(Table):
|
|||||||
or raise an error if no corresponding embedding function is found.
|
or raise an error if no corresponding embedding function is found.
|
||||||
If the `query` is a string, then the query type is "vector" if the
|
If the `query` is a string, then the query type is "vector" if the
|
||||||
table has embedding functions, else the query type is "fts"
|
table has embedding functions, else the query type is "fts"
|
||||||
|
fts_columns: str or list of str, default None
|
||||||
|
The column(s) to search in for full-text search.
|
||||||
|
If None then the search is performed on all indexed columns.
|
||||||
|
For now, only one column can be searched at a time.
|
||||||
|
|
||||||
Returns
|
Returns
|
||||||
-------
|
-------
|
||||||
@@ -1547,7 +1572,7 @@ class LanceTable(Table):
|
|||||||
metadata = registry.get_table_metadata(embedding_functions)
|
metadata = registry.get_table_metadata(embedding_functions)
|
||||||
|
|
||||||
if data is not None:
|
if data is not None:
|
||||||
data = _sanitize_data(
|
data, schema = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
schema,
|
schema,
|
||||||
metadata=metadata,
|
metadata=metadata,
|
||||||
@@ -1664,6 +1689,7 @@ class LanceTable(Table):
|
|||||||
"nprobes": query.nprobes,
|
"nprobes": query.nprobes,
|
||||||
"refine_factor": query.refine_factor,
|
"refine_factor": query.refine_factor,
|
||||||
},
|
},
|
||||||
|
full_text_query=query.full_text_query,
|
||||||
with_row_id=query.with_row_id,
|
with_row_id=query.with_row_id,
|
||||||
batch_size=batch_size,
|
batch_size=batch_size,
|
||||||
).to_reader()
|
).to_reader()
|
||||||
@@ -1675,7 +1701,7 @@ class LanceTable(Table):
|
|||||||
on_bad_vectors: str,
|
on_bad_vectors: str,
|
||||||
fill_value: float,
|
fill_value: float,
|
||||||
):
|
):
|
||||||
new_data = _sanitize_data(
|
new_data, _ = _sanitize_data(
|
||||||
new_data,
|
new_data,
|
||||||
self.schema,
|
self.schema,
|
||||||
metadata=self.schema.metadata,
|
metadata=self.schema.metadata,
|
||||||
@@ -2087,7 +2113,7 @@ class AsyncTable:
|
|||||||
column: str,
|
column: str,
|
||||||
*,
|
*,
|
||||||
replace: Optional[bool] = None,
|
replace: Optional[bool] = None,
|
||||||
config: Optional[Union[IvfPq, BTree]] = None,
|
config: Optional[Union[IvfPq, BTree, Bitmap, LabelList]] = None,
|
||||||
):
|
):
|
||||||
"""Create an index to speed up queries
|
"""Create an index to speed up queries
|
||||||
|
|
||||||
@@ -2153,7 +2179,7 @@ class AsyncTable:
|
|||||||
on_bad_vectors = "error"
|
on_bad_vectors = "error"
|
||||||
if fill_value is None:
|
if fill_value is None:
|
||||||
fill_value = 0.0
|
fill_value = 0.0
|
||||||
data = _sanitize_data(
|
data, _ = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
schema,
|
schema,
|
||||||
metadata=schema.metadata,
|
metadata=schema.metadata,
|
||||||
|
|||||||
@@ -22,7 +22,8 @@ import pytest
|
|||||||
from lancedb.pydantic import LanceModel, Vector
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
|
||||||
|
|
||||||
def test_basic(tmp_path):
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_basic(tmp_path, use_tantivy):
|
||||||
db = lancedb.connect(tmp_path)
|
db = lancedb.connect(tmp_path)
|
||||||
|
|
||||||
assert db.uri == str(tmp_path)
|
assert db.uri == str(tmp_path)
|
||||||
@@ -55,7 +56,7 @@ def test_basic(tmp_path):
|
|||||||
assert len(rs) == 1
|
assert len(rs) == 1
|
||||||
assert rs["item"].iloc[0] == "foo"
|
assert rs["item"].iloc[0] == "foo"
|
||||||
|
|
||||||
table.create_fts_index(["item"])
|
table.create_fts_index("item", use_tantivy=use_tantivy)
|
||||||
rs = table.search("bar", query_type="fts").to_pandas()
|
rs = table.search("bar", query_type="fts").to_pandas()
|
||||||
assert len(rs) == 1
|
assert len(rs) == 1
|
||||||
assert rs["item"].iloc[0] == "bar"
|
assert rs["item"].iloc[0] == "bar"
|
||||||
|
|||||||
@@ -417,3 +417,28 @@ def test_openai_embedding(tmp_path):
|
|||||||
tbl.add(df)
|
tbl.add(df)
|
||||||
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
||||||
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.slow
|
||||||
|
@pytest.mark.skipif(
|
||||||
|
os.environ.get("WATSONX_API_KEY") is None
|
||||||
|
or os.environ.get("WATSONX_PROJECT_ID") is None,
|
||||||
|
reason="WATSONX_API_KEY and WATSONX_PROJECT_ID not set",
|
||||||
|
)
|
||||||
|
def test_watsonx_embedding(tmp_path):
|
||||||
|
from lancedb.embeddings import WatsonxEmbeddings
|
||||||
|
|
||||||
|
for name in WatsonxEmbeddings.model_names():
|
||||||
|
model = get_registry().get("watsonx").create(max_retries=0, name=name)
|
||||||
|
|
||||||
|
class TextModel(LanceModel):
|
||||||
|
text: str = model.SourceField()
|
||||||
|
vector: Vector(model.ndims()) = model.VectorField()
|
||||||
|
|
||||||
|
db = lancedb.connect("~/.lancedb")
|
||||||
|
tbl = db.create_table("watsonx_test", schema=TextModel, mode="overwrite")
|
||||||
|
df = pd.DataFrame({"text": ["hello world", "goodbye world"]})
|
||||||
|
|
||||||
|
tbl.add(df)
|
||||||
|
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
||||||
|
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
||||||
|
|||||||
@@ -74,7 +74,12 @@ def test_create_index_with_stemming(tmp_path, table):
|
|||||||
assert os.path.exists(str(tmp_path / "index"))
|
assert os.path.exists(str(tmp_path / "index"))
|
||||||
|
|
||||||
# Check stemming by running tokenizer on non empty table
|
# Check stemming by running tokenizer on non empty table
|
||||||
table.create_fts_index("text", tokenizer_name="en_stem")
|
table.create_fts_index("text", tokenizer_name="en_stem", use_tantivy=True)
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_create_inverted_index(table, use_tantivy):
|
||||||
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
|
|
||||||
|
|
||||||
def test_populate_index(tmp_path, table):
|
def test_populate_index(tmp_path, table):
|
||||||
@@ -92,8 +97,15 @@ def test_search_index(tmp_path, table):
|
|||||||
assert len(results[1]) == 10 # _distance
|
assert len(results[1]) == 10 # _distance
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_search_fts(table, use_tantivy):
|
||||||
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
|
results = table.search("puppy").limit(10).to_list()
|
||||||
|
assert len(results) == 10
|
||||||
|
|
||||||
|
|
||||||
def test_search_ordering_field_index_table(tmp_path, table):
|
def test_search_ordering_field_index_table(tmp_path, table):
|
||||||
table.create_fts_index("text", ordering_field_names=["count"])
|
table.create_fts_index("text", ordering_field_names=["count"], use_tantivy=True)
|
||||||
rows = (
|
rows = (
|
||||||
table.search("puppy", ordering_field_name="count")
|
table.search("puppy", ordering_field_name="count")
|
||||||
.limit(20)
|
.limit(20)
|
||||||
@@ -125,8 +137,9 @@ def test_search_ordering_field_index(tmp_path, table):
|
|||||||
assert sorted(rows, key=lambda x: x["count"], reverse=True) == rows
|
assert sorted(rows, key=lambda x: x["count"], reverse=True) == rows
|
||||||
|
|
||||||
|
|
||||||
def test_create_index_from_table(tmp_path, table):
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
table.create_fts_index("text")
|
def test_create_index_from_table(tmp_path, table, use_tantivy):
|
||||||
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
df = table.search("puppy").limit(10).select(["text"]).to_pandas()
|
df = table.search("puppy").limit(10).select(["text"]).to_pandas()
|
||||||
assert len(df) <= 10
|
assert len(df) <= 10
|
||||||
assert "text" in df.columns
|
assert "text" in df.columns
|
||||||
@@ -145,15 +158,15 @@ def test_create_index_from_table(tmp_path, table):
|
|||||||
]
|
]
|
||||||
)
|
)
|
||||||
|
|
||||||
with pytest.raises(ValueError, match="already exists"):
|
with pytest.raises(Exception, match="already exists"):
|
||||||
table.create_fts_index("text")
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
|
|
||||||
table.create_fts_index("text", replace=True)
|
table.create_fts_index("text", replace=True, use_tantivy=use_tantivy)
|
||||||
assert len(table.search("gorilla").limit(1).to_pandas()) == 1
|
assert len(table.search("gorilla").limit(1).to_pandas()) == 1
|
||||||
|
|
||||||
|
|
||||||
def test_create_index_multiple_columns(tmp_path, table):
|
def test_create_index_multiple_columns(tmp_path, table):
|
||||||
table.create_fts_index(["text", "text2"])
|
table.create_fts_index(["text", "text2"], use_tantivy=True)
|
||||||
df = table.search("puppy").limit(10).to_pandas()
|
df = table.search("puppy").limit(10).to_pandas()
|
||||||
assert len(df) == 10
|
assert len(df) == 10
|
||||||
assert "text" in df.columns
|
assert "text" in df.columns
|
||||||
@@ -161,20 +174,21 @@ def test_create_index_multiple_columns(tmp_path, table):
|
|||||||
|
|
||||||
|
|
||||||
def test_empty_rs(tmp_path, table, mocker):
|
def test_empty_rs(tmp_path, table, mocker):
|
||||||
table.create_fts_index(["text", "text2"])
|
table.create_fts_index(["text", "text2"], use_tantivy=True)
|
||||||
mocker.patch("lancedb.fts.search_index", return_value=([], []))
|
mocker.patch("lancedb.fts.search_index", return_value=([], []))
|
||||||
df = table.search("puppy").limit(10).to_pandas()
|
df = table.search("puppy").limit(10).to_pandas()
|
||||||
assert len(df) == 0
|
assert len(df) == 0
|
||||||
|
|
||||||
|
|
||||||
def test_nested_schema(tmp_path, table):
|
def test_nested_schema(tmp_path, table):
|
||||||
table.create_fts_index("nested.text")
|
table.create_fts_index("nested.text", use_tantivy=True)
|
||||||
rs = table.search("puppy").limit(10).to_list()
|
rs = table.search("puppy").limit(10).to_list()
|
||||||
assert len(rs) == 10
|
assert len(rs) == 10
|
||||||
|
|
||||||
|
|
||||||
def test_search_index_with_filter(table):
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
table.create_fts_index("text")
|
def test_search_index_with_filter(table, use_tantivy):
|
||||||
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
orig_import = __import__
|
orig_import = __import__
|
||||||
|
|
||||||
def import_mock(name, *args):
|
def import_mock(name, *args):
|
||||||
@@ -186,7 +200,7 @@ def test_search_index_with_filter(table):
|
|||||||
with mock.patch("builtins.__import__", side_effect=import_mock):
|
with mock.patch("builtins.__import__", side_effect=import_mock):
|
||||||
rs = table.search("puppy").where("id=1").limit(10)
|
rs = table.search("puppy").where("id=1").limit(10)
|
||||||
# test schema
|
# test schema
|
||||||
assert rs.to_arrow().drop("score").schema.equals(table.schema)
|
assert rs.to_arrow().drop("_score").schema.equals(table.schema)
|
||||||
|
|
||||||
rs = rs.to_list()
|
rs = rs.to_list()
|
||||||
for r in rs:
|
for r in rs:
|
||||||
@@ -204,7 +218,8 @@ def test_search_index_with_filter(table):
|
|||||||
assert r["_rowid"] is not None
|
assert r["_rowid"] is not None
|
||||||
|
|
||||||
|
|
||||||
def test_null_input(table):
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_null_input(table, use_tantivy):
|
||||||
table.add(
|
table.add(
|
||||||
[
|
[
|
||||||
{
|
{
|
||||||
@@ -217,12 +232,12 @@ def test_null_input(table):
|
|||||||
}
|
}
|
||||||
]
|
]
|
||||||
)
|
)
|
||||||
table.create_fts_index("text")
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
|
|
||||||
|
|
||||||
def test_syntax(table):
|
def test_syntax(table):
|
||||||
# https://github.com/lancedb/lancedb/issues/769
|
# https://github.com/lancedb/lancedb/issues/769
|
||||||
table.create_fts_index("text")
|
table.create_fts_index("text", use_tantivy=True)
|
||||||
with pytest.raises(ValueError, match="Syntax Error"):
|
with pytest.raises(ValueError, match="Syntax Error"):
|
||||||
table.search("they could have been dogs OR").limit(10).to_list()
|
table.search("they could have been dogs OR").limit(10).to_list()
|
||||||
|
|
||||||
|
|||||||
@@ -124,3 +124,17 @@ def test_bad_hf_dataset(tmp_path: Path, mock_embedding_function, hf_dataset_with
|
|||||||
# this should still work because we don't add the split column
|
# this should still work because we don't add the split column
|
||||||
# if it already exists
|
# if it already exists
|
||||||
train_table.add(hf_dataset_with_split)
|
train_table.add(hf_dataset_with_split)
|
||||||
|
|
||||||
|
|
||||||
|
def test_generator(tmp_path: Path):
|
||||||
|
db = lancedb.connect(tmp_path)
|
||||||
|
|
||||||
|
def gen():
|
||||||
|
yield {"pokemon": "bulbasaur", "type": "grass"}
|
||||||
|
yield {"pokemon": "squirtle", "type": "water"}
|
||||||
|
|
||||||
|
ds = datasets.Dataset.from_generator(gen)
|
||||||
|
tbl = db.create_table("pokemon", ds)
|
||||||
|
|
||||||
|
assert len(tbl) == 2
|
||||||
|
assert tbl.schema == ds.features.arrow_schema
|
||||||
|
|||||||
@@ -1,10 +1,14 @@
|
|||||||
|
# SPDX-License-Identifier: Apache-2.0
|
||||||
|
# SPDX-FileCopyrightText: Copyright The LanceDB Authors
|
||||||
|
|
||||||
from datetime import timedelta
|
from datetime import timedelta
|
||||||
|
import random
|
||||||
|
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
import pytest
|
import pytest
|
||||||
import pytest_asyncio
|
import pytest_asyncio
|
||||||
from lancedb import AsyncConnection, AsyncTable, connect_async
|
from lancedb import AsyncConnection, AsyncTable, connect_async
|
||||||
from lancedb.index import BTree, IvfPq
|
from lancedb.index import BTree, IvfPq, Bitmap, LabelList
|
||||||
|
|
||||||
|
|
||||||
@pytest_asyncio.fixture
|
@pytest_asyncio.fixture
|
||||||
@@ -25,8 +29,11 @@ NROWS = 256
|
|||||||
async def some_table(db_async):
|
async def some_table(db_async):
|
||||||
data = pa.Table.from_pydict(
|
data = pa.Table.from_pydict(
|
||||||
{
|
{
|
||||||
"id": list(range(256)),
|
"id": list(range(NROWS)),
|
||||||
"vector": sample_fixed_size_list_array(NROWS, DIM),
|
"vector": sample_fixed_size_list_array(NROWS, DIM),
|
||||||
|
"tags": [
|
||||||
|
[f"tag{random.randint(0, 8)}" for _ in range(2)] for _ in range(NROWS)
|
||||||
|
],
|
||||||
}
|
}
|
||||||
)
|
)
|
||||||
return await db_async.create_table(
|
return await db_async.create_table(
|
||||||
@@ -42,6 +49,7 @@ async def test_create_scalar_index(some_table: AsyncTable):
|
|||||||
# Can recreate if replace=True
|
# Can recreate if replace=True
|
||||||
await some_table.create_index("id", replace=True)
|
await some_table.create_index("id", replace=True)
|
||||||
indices = await some_table.list_indices()
|
indices = await some_table.list_indices()
|
||||||
|
assert str(indices) == '[Index(BTree, columns=["id"])]'
|
||||||
assert len(indices) == 1
|
assert len(indices) == 1
|
||||||
assert indices[0].index_type == "BTree"
|
assert indices[0].index_type == "BTree"
|
||||||
assert indices[0].columns == ["id"]
|
assert indices[0].columns == ["id"]
|
||||||
@@ -52,6 +60,22 @@ async def test_create_scalar_index(some_table: AsyncTable):
|
|||||||
await some_table.create_index("id", config=BTree())
|
await some_table.create_index("id", config=BTree())
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.asyncio
|
||||||
|
async def test_create_bitmap_index(some_table: AsyncTable):
|
||||||
|
await some_table.create_index("id", config=Bitmap())
|
||||||
|
# TODO: Fix via https://github.com/lancedb/lance/issues/2039
|
||||||
|
# indices = await some_table.list_indices()
|
||||||
|
# assert str(indices) == '[Index(Bitmap, columns=["id"])]'
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.asyncio
|
||||||
|
async def test_create_label_list_index(some_table: AsyncTable):
|
||||||
|
await some_table.create_index("tags", config=LabelList())
|
||||||
|
# TODO: Fix via https://github.com/lancedb/lance/issues/2039
|
||||||
|
# indices = await some_table.list_indices()
|
||||||
|
# assert str(indices) == '[Index(LabelList, columns=["id"])]'
|
||||||
|
|
||||||
|
|
||||||
@pytest.mark.asyncio
|
@pytest.mark.asyncio
|
||||||
async def test_create_vector_index(some_table: AsyncTable):
|
async def test_create_vector_index(some_table: AsyncTable):
|
||||||
# Can create
|
# Can create
|
||||||
|
|||||||
@@ -354,3 +354,11 @@ async def test_query_camelcase_async(tmp_path):
|
|||||||
|
|
||||||
result = await table.query().select(["camelCase"]).to_arrow()
|
result = await table.query().select(["camelCase"]).to_arrow()
|
||||||
assert result == pa.table({"camelCase": pa.array([1, 2])})
|
assert result == pa.table({"camelCase": pa.array([1, 2])})
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.asyncio
|
||||||
|
async def test_query_to_list_async(table_async: AsyncTable):
|
||||||
|
list = await table_async.query().to_list()
|
||||||
|
assert len(list) == 2
|
||||||
|
assert list[0]["vector"] == [1, 2]
|
||||||
|
assert list[1]["vector"] == [3, 4]
|
||||||
|
|||||||
@@ -1,4 +1,5 @@
|
|||||||
import os
|
import os
|
||||||
|
import random
|
||||||
|
|
||||||
import lancedb
|
import lancedb
|
||||||
import numpy as np
|
import numpy as np
|
||||||
@@ -7,6 +8,8 @@ from lancedb.conftest import MockTextEmbeddingFunction # noqa
|
|||||||
from lancedb.embeddings import EmbeddingFunctionRegistry
|
from lancedb.embeddings import EmbeddingFunctionRegistry
|
||||||
from lancedb.pydantic import LanceModel, Vector
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
from lancedb.rerankers import (
|
from lancedb.rerankers import (
|
||||||
|
LinearCombinationReranker,
|
||||||
|
RRFReranker,
|
||||||
CohereReranker,
|
CohereReranker,
|
||||||
ColbertReranker,
|
ColbertReranker,
|
||||||
CrossEncoderReranker,
|
CrossEncoderReranker,
|
||||||
@@ -19,14 +22,17 @@ from lancedb.table import LanceTable
|
|||||||
pytest.importorskip("lancedb.fts")
|
pytest.importorskip("lancedb.fts")
|
||||||
|
|
||||||
|
|
||||||
def get_test_table(tmp_path):
|
def get_test_table(tmp_path, use_tantivy):
|
||||||
db = lancedb.connect(tmp_path)
|
db = lancedb.connect(tmp_path)
|
||||||
# Create a LanceDB table schema with a vector and a text column
|
# Create a LanceDB table schema with a vector and a text column
|
||||||
emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
||||||
|
meta_emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
||||||
|
|
||||||
class MyTable(LanceModel):
|
class MyTable(LanceModel):
|
||||||
text: str = emb.SourceField()
|
text: str = emb.SourceField()
|
||||||
vector: Vector(emb.ndims()) = emb.VectorField()
|
vector: Vector(emb.ndims()) = emb.VectorField()
|
||||||
|
meta: str = meta_emb.SourceField()
|
||||||
|
meta_vector: Vector(meta_emb.ndims()) = meta_emb.VectorField()
|
||||||
|
|
||||||
# Initialize the table using the schema
|
# Initialize the table using the schema
|
||||||
table = LanceTable.create(
|
table = LanceTable.create(
|
||||||
@@ -75,10 +81,15 @@ def get_test_table(tmp_path):
|
|||||||
]
|
]
|
||||||
|
|
||||||
# Add the phrases and vectors to the table
|
# Add the phrases and vectors to the table
|
||||||
table.add([{"text": p} for p in phrases])
|
table.add(
|
||||||
|
[
|
||||||
|
{"text": p, "meta": phrases[random.randint(0, len(phrases) - 1)]}
|
||||||
|
for p in phrases
|
||||||
|
]
|
||||||
|
)
|
||||||
|
|
||||||
# Create a fts index
|
# Create a fts index
|
||||||
table.create_fts_index("text")
|
table.create_fts_index("text", use_tantivy=use_tantivy)
|
||||||
|
|
||||||
return table, MyTable
|
return table, MyTable
|
||||||
|
|
||||||
@@ -86,12 +97,12 @@ def get_test_table(tmp_path):
|
|||||||
def _run_test_reranker(reranker, table, query, query_vector, schema):
|
def _run_test_reranker(reranker, table, query, query_vector, schema):
|
||||||
# Hybrid search setting
|
# Hybrid search setting
|
||||||
result1 = (
|
result1 = (
|
||||||
table.search(query, query_type="hybrid")
|
table.search(query, query_type="hybrid", vector_column_name="vector")
|
||||||
.rerank(normalize="score", reranker=reranker)
|
.rerank(normalize="score", reranker=reranker)
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result2 = (
|
result2 = (
|
||||||
table.search(query, query_type="hybrid")
|
table.search(query, query_type="hybrid", vector_column_name="vector")
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
@@ -99,7 +110,7 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
|
|
||||||
query_vector = table.to_pandas()["vector"][0]
|
query_vector = table.to_pandas()["vector"][0]
|
||||||
result = (
|
result = (
|
||||||
table.search((query_vector, query))
|
table.search((query_vector, query), vector_column_name="vector")
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -114,11 +125,16 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
|
|
||||||
# Vector search setting
|
# Vector search setting
|
||||||
result = table.search(query).rerank(reranker=reranker).limit(30).to_arrow()
|
result = (
|
||||||
|
table.search(query, vector_column_name="vector")
|
||||||
|
.rerank(reranker=reranker)
|
||||||
|
.limit(30)
|
||||||
|
.to_arrow()
|
||||||
|
)
|
||||||
assert len(result) == 30
|
assert len(result) == 30
|
||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
result_explicit = (
|
result_explicit = (
|
||||||
table.search(query_vector)
|
table.search(query_vector, vector_column_name="vector")
|
||||||
.rerank(reranker=reranker, query_string=query)
|
.rerank(reranker=reranker, query_string=query)
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -127,11 +143,13 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
with pytest.raises(
|
with pytest.raises(
|
||||||
ValueError
|
ValueError
|
||||||
): # This raises an error because vector query is provided without reanking query
|
): # This raises an error because vector query is provided without reanking query
|
||||||
table.search(query_vector).rerank(reranker=reranker).limit(30).to_arrow()
|
table.search(query_vector, vector_column_name="vector").rerank(
|
||||||
|
reranker=reranker
|
||||||
|
).limit(30).to_arrow()
|
||||||
|
|
||||||
# FTS search setting
|
# FTS search setting
|
||||||
result = (
|
result = (
|
||||||
table.search(query, query_type="fts")
|
table.search(query, query_type="fts", vector_column_name="vector")
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -139,22 +157,48 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
assert len(result) > 0
|
assert len(result) > 0
|
||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
|
|
||||||
|
# Multi-vector search setting
|
||||||
|
rs1 = table.search(query, vector_column_name="vector").limit(10).with_row_id(True)
|
||||||
|
rs2 = (
|
||||||
|
table.search(query, vector_column_name="meta_vector")
|
||||||
|
.limit(10)
|
||||||
|
.with_row_id(True)
|
||||||
|
)
|
||||||
|
result = reranker.rerank_multivector([rs1, rs2], query)
|
||||||
|
assert len(result) == 20
|
||||||
|
result_deduped = reranker.rerank_multivector(
|
||||||
|
[rs1, rs2, rs1], query, deduplicate=True
|
||||||
|
)
|
||||||
|
assert len(result_deduped) < 20
|
||||||
|
result_arrow = reranker.rerank_multivector([rs1.to_arrow(), rs2.to_arrow()], query)
|
||||||
|
assert len(result) == 20 and result == result_arrow
|
||||||
|
|
||||||
def test_linear_combination(tmp_path):
|
|
||||||
table, schema = get_test_table(tmp_path)
|
def _run_test_hybrid_reranker(reranker, tmp_path, use_tantivy):
|
||||||
|
table, schema = get_test_table(tmp_path, use_tantivy)
|
||||||
# The default reranker
|
# The default reranker
|
||||||
result1 = (
|
result1 = (
|
||||||
table.search("Our father who art in heaven", query_type="hybrid")
|
table.search(
|
||||||
|
"Our father who art in heaven",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
|
)
|
||||||
.rerank(normalize="score")
|
.rerank(normalize="score")
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result2 = ( # noqa
|
result2 = ( # noqa
|
||||||
table.search("Our father who art in heaven.", query_type="hybrid")
|
table.search(
|
||||||
|
"Our father who art in heaven.",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
|
)
|
||||||
.rerank(normalize="rank")
|
.rerank(normalize="rank")
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result3 = table.search(
|
result3 = table.search(
|
||||||
"Our father who art in heaven..", query_type="hybrid"
|
"Our father who art in heaven..",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
).to_pydantic(schema)
|
).to_pydantic(schema)
|
||||||
|
|
||||||
assert result1 == result3 # 2 & 3 should be the same as they use score as score
|
assert result1 == result3 # 2 & 3 should be the same as they use score as score
|
||||||
@@ -162,7 +206,7 @@ def test_linear_combination(tmp_path):
|
|||||||
query = "Our father who art in heaven"
|
query = "Our father who art in heaven"
|
||||||
query_vector = table.to_pandas()["vector"][0]
|
query_vector = table.to_pandas()["vector"][0]
|
||||||
result = (
|
result = (
|
||||||
table.search((query_vector, query))
|
table.search((query_vector, query), vector_column_name="vector")
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.rerank(normalize="score")
|
.rerank(normalize="score")
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -177,6 +221,18 @@ def test_linear_combination(tmp_path):
|
|||||||
)
|
)
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_linear_combination(tmp_path, use_tantivy):
|
||||||
|
reranker = LinearCombinationReranker()
|
||||||
|
_run_test_hybrid_reranker(reranker, tmp_path, use_tantivy)
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.parametrize("use_tantivy", [True, False])
|
||||||
|
def test_rrf_reranker(tmp_path, use_tantivy):
|
||||||
|
reranker = RRFReranker()
|
||||||
|
_run_test_hybrid_reranker(reranker, tmp_path, use_tantivy)
|
||||||
|
|
||||||
|
|
||||||
@pytest.mark.skipif(
|
@pytest.mark.skipif(
|
||||||
os.environ.get("COHERE_API_KEY") is None, reason="COHERE_API_KEY not set"
|
os.environ.get("COHERE_API_KEY") is None, reason="COHERE_API_KEY not set"
|
||||||
)
|
)
|
||||||
|
|||||||
@@ -730,7 +730,7 @@ def test_create_scalar_index(db):
|
|||||||
indices = table.to_lance().list_indices()
|
indices = table.to_lance().list_indices()
|
||||||
assert len(indices) == 1
|
assert len(indices) == 1
|
||||||
scalar_index = indices[0]
|
scalar_index = indices[0]
|
||||||
assert scalar_index["type"] == "Scalar"
|
assert scalar_index["type"] == "BTree"
|
||||||
|
|
||||||
# Confirm that prefiltering still works with the scalar index column
|
# Confirm that prefiltering still works with the scalar index column
|
||||||
results = table.search().where("x = 'c'").to_arrow()
|
results = table.search().where("x = 'c'").to_arrow()
|
||||||
@@ -1034,6 +1034,12 @@ async def test_optimize(db_async: AsyncConnection):
|
|||||||
],
|
],
|
||||||
)
|
)
|
||||||
stats = await table.optimize()
|
stats = await table.optimize()
|
||||||
|
expected = (
|
||||||
|
"OptimizeStats(compaction=CompactionStats { fragments_removed: 2, "
|
||||||
|
"fragments_added: 1, files_removed: 2, files_added: 1 }, "
|
||||||
|
"prune=RemovalStats { bytes_removed: 0, old_versions_removed: 0 })"
|
||||||
|
)
|
||||||
|
assert str(stats) == expected
|
||||||
assert stats.compaction.files_removed == 2
|
assert stats.compaction.files_removed == 2
|
||||||
assert stats.compaction.files_added == 1
|
assert stats.compaction.files_added == 1
|
||||||
assert stats.compaction.fragments_added == 1
|
assert stats.compaction.fragments_added == 1
|
||||||
|
|||||||
Some files were not shown because too many files have changed in this diff Show More
Reference in New Issue
Block a user