mirror of
https://github.com/lancedb/lancedb.git
synced 2025-12-23 21:39:57 +00:00
Compare commits
20 Commits
python-v0.
...
python-v0.
| Author | SHA1 | Date | |
|---|---|---|---|
|
|
ec39d98571 | ||
|
|
0cb37f0e5e | ||
|
|
24e3507ee2 | ||
|
|
2bdf0a02f9 | ||
|
|
32123713fd | ||
|
|
d5a01ffe7b | ||
|
|
e01045692c | ||
|
|
a62f661d90 | ||
|
|
4769d8eb76 | ||
|
|
d07d7a5980 | ||
|
|
8d2ff7b210 | ||
|
|
61c05b51a0 | ||
|
|
7801ab9b8b | ||
|
|
d297da5a7e | ||
|
|
6af69b57ad | ||
|
|
a062a92f6b | ||
|
|
277b753fd8 | ||
|
|
f78b7863f6 | ||
|
|
e7d824af2b | ||
|
|
02f1ec775f |
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.7.2"
|
current_version = "0.8.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
|
|||||||
48
.github/workflows/java.yml
vendored
48
.github/workflows/java.yml
vendored
@@ -3,6 +3,8 @@ on:
|
|||||||
push:
|
push:
|
||||||
branches:
|
branches:
|
||||||
- main
|
- main
|
||||||
|
paths:
|
||||||
|
- java/**
|
||||||
pull_request:
|
pull_request:
|
||||||
paths:
|
paths:
|
||||||
- java/**
|
- java/**
|
||||||
@@ -21,9 +23,42 @@ env:
|
|||||||
CARGO_INCREMENTAL: "0"
|
CARGO_INCREMENTAL: "0"
|
||||||
CARGO_BUILD_JOBS: "1"
|
CARGO_BUILD_JOBS: "1"
|
||||||
jobs:
|
jobs:
|
||||||
linux-build:
|
linux-build-java-11:
|
||||||
runs-on: ubuntu-22.04
|
runs-on: ubuntu-22.04
|
||||||
name: ubuntu-22.04 + Java 11 & 17
|
name: ubuntu-22.04 + Java 11
|
||||||
|
defaults:
|
||||||
|
run:
|
||||||
|
working-directory: ./java
|
||||||
|
steps:
|
||||||
|
- name: Checkout repository
|
||||||
|
uses: actions/checkout@v4
|
||||||
|
- uses: Swatinem/rust-cache@v2
|
||||||
|
with:
|
||||||
|
workspaces: java/core/lancedb-jni
|
||||||
|
- name: Run cargo fmt
|
||||||
|
run: cargo fmt --check
|
||||||
|
working-directory: ./java/core/lancedb-jni
|
||||||
|
- name: Install dependencies
|
||||||
|
run: |
|
||||||
|
sudo apt update
|
||||||
|
sudo apt install -y protobuf-compiler libssl-dev
|
||||||
|
- name: Install Java 11
|
||||||
|
uses: actions/setup-java@v4
|
||||||
|
with:
|
||||||
|
distribution: temurin
|
||||||
|
java-version: 11
|
||||||
|
cache: "maven"
|
||||||
|
- name: Java Style Check
|
||||||
|
run: mvn checkstyle:check
|
||||||
|
# Disable because of issues in lancedb rust core code
|
||||||
|
# - name: Rust Clippy
|
||||||
|
# working-directory: java/core/lancedb-jni
|
||||||
|
# run: cargo clippy --all-targets -- -D warnings
|
||||||
|
- name: Running tests with Java 11
|
||||||
|
run: mvn clean test
|
||||||
|
linux-build-java-17:
|
||||||
|
runs-on: ubuntu-22.04
|
||||||
|
name: ubuntu-22.04 + Java 17
|
||||||
defaults:
|
defaults:
|
||||||
run:
|
run:
|
||||||
working-directory: ./java
|
working-directory: ./java
|
||||||
@@ -47,20 +82,12 @@ jobs:
|
|||||||
java-version: 17
|
java-version: 17
|
||||||
cache: "maven"
|
cache: "maven"
|
||||||
- run: echo "JAVA_17=$JAVA_HOME" >> $GITHUB_ENV
|
- run: echo "JAVA_17=$JAVA_HOME" >> $GITHUB_ENV
|
||||||
- name: Install Java 11
|
|
||||||
uses: actions/setup-java@v4
|
|
||||||
with:
|
|
||||||
distribution: temurin
|
|
||||||
java-version: 11
|
|
||||||
cache: "maven"
|
|
||||||
- name: Java Style Check
|
- name: Java Style Check
|
||||||
run: mvn checkstyle:check
|
run: mvn checkstyle:check
|
||||||
# Disable because of issues in lancedb rust core code
|
# Disable because of issues in lancedb rust core code
|
||||||
# - name: Rust Clippy
|
# - name: Rust Clippy
|
||||||
# working-directory: java/core/lancedb-jni
|
# working-directory: java/core/lancedb-jni
|
||||||
# run: cargo clippy --all-targets -- -D warnings
|
# run: cargo clippy --all-targets -- -D warnings
|
||||||
- name: Running tests with Java 11
|
|
||||||
run: mvn clean test
|
|
||||||
- name: Running tests with Java 17
|
- name: Running tests with Java 17
|
||||||
run: |
|
run: |
|
||||||
export JAVA_TOOL_OPTIONS="$JAVA_TOOL_OPTIONS \
|
export JAVA_TOOL_OPTIONS="$JAVA_TOOL_OPTIONS \
|
||||||
@@ -83,3 +110,4 @@ jobs:
|
|||||||
-Djdk.reflect.useDirectMethodHandle=false \
|
-Djdk.reflect.useDirectMethodHandle=false \
|
||||||
-Dio.netty.tryReflectionSetAccessible=true"
|
-Dio.netty.tryReflectionSetAccessible=true"
|
||||||
JAVA_HOME=$JAVA_17 mvn clean test
|
JAVA_HOME=$JAVA_17 mvn clean test
|
||||||
|
|
||||||
|
|||||||
27
Cargo.toml
27
Cargo.toml
@@ -20,20 +20,21 @@ keywords = ["lancedb", "lance", "database", "vector", "search"]
|
|||||||
categories = ["database-implementations"]
|
categories = ["database-implementations"]
|
||||||
|
|
||||||
[workspace.dependencies]
|
[workspace.dependencies]
|
||||||
lance = { "version" = "=0.15.0", "features" = ["dynamodb"] }
|
lance = { "version" = "=0.16.0", "features" = ["dynamodb"] }
|
||||||
lance-index = { "version" = "=0.15.0" }
|
lance-index = { "version" = "=0.16.0" }
|
||||||
lance-linalg = { "version" = "=0.15.0" }
|
lance-linalg = { "version" = "=0.16.0" }
|
||||||
lance-testing = { "version" = "=0.15.0" }
|
lance-testing = { "version" = "=0.16.0" }
|
||||||
lance-datafusion = { "version" = "=0.15.0" }
|
lance-datafusion = { "version" = "=0.16.0" }
|
||||||
|
lance-encoding = { "version" = "=0.16.0" }
|
||||||
# Note that this one does not include pyarrow
|
# Note that this one does not include pyarrow
|
||||||
arrow = { version = "52.1", optional = false }
|
arrow = { version = "52.2", optional = false }
|
||||||
arrow-array = "52.1"
|
arrow-array = "52.2"
|
||||||
arrow-data = "52.1"
|
arrow-data = "52.2"
|
||||||
arrow-ipc = "52.1"
|
arrow-ipc = "52.2"
|
||||||
arrow-ord = "52.1"
|
arrow-ord = "52.2"
|
||||||
arrow-schema = "52.1"
|
arrow-schema = "52.2"
|
||||||
arrow-arith = "52.1"
|
arrow-arith = "52.2"
|
||||||
arrow-cast = "52.1"
|
arrow-cast = "52.2"
|
||||||
async-trait = "0"
|
async-trait = "0"
|
||||||
chrono = "0.4.35"
|
chrono = "0.4.35"
|
||||||
datafusion-physical-plan = "40.0"
|
datafusion-physical-plan = "40.0"
|
||||||
|
|||||||
@@ -18,4 +18,4 @@ docker run \
|
|||||||
-v $(pwd):/io -w /io \
|
-v $(pwd):/io -w /io \
|
||||||
--memory-swap=-1 \
|
--memory-swap=-1 \
|
||||||
lancedb-node-manylinux \
|
lancedb-node-manylinux \
|
||||||
bash ci/manylinux_node/build.sh $ARCH
|
bash ci/manylinux_node/build_vectordb.sh $ARCH
|
||||||
|
|||||||
@@ -4,9 +4,9 @@ ARCH=${1:-x86_64}
|
|||||||
|
|
||||||
# We pass down the current user so that when we later mount the local files
|
# We pass down the current user so that when we later mount the local files
|
||||||
# into the container, the files are accessible by the current user.
|
# into the container, the files are accessible by the current user.
|
||||||
pushd ci/manylinux_nodejs
|
pushd ci/manylinux_node
|
||||||
docker build \
|
docker build \
|
||||||
-t lancedb-nodejs-manylinux \
|
-t lancedb-node-manylinux-$ARCH \
|
||||||
--build-arg="ARCH=$ARCH" \
|
--build-arg="ARCH=$ARCH" \
|
||||||
--build-arg="DOCKER_USER=$(id -u)" \
|
--build-arg="DOCKER_USER=$(id -u)" \
|
||||||
--progress=plain \
|
--progress=plain \
|
||||||
@@ -17,5 +17,5 @@ popd
|
|||||||
docker run \
|
docker run \
|
||||||
-v $(pwd):/io -w /io \
|
-v $(pwd):/io -w /io \
|
||||||
--memory-swap=-1 \
|
--memory-swap=-1 \
|
||||||
lancedb-nodejs-manylinux \
|
lancedb-node-manylinux-$ARCH \
|
||||||
bash ci/manylinux_nodejs/build.sh $ARCH
|
bash ci/manylinux_node/build_lancedb.sh $ARCH
|
||||||
|
|||||||
@@ -4,7 +4,7 @@
|
|||||||
# range of linux distributions.
|
# range of linux distributions.
|
||||||
ARG ARCH=x86_64
|
ARG ARCH=x86_64
|
||||||
|
|
||||||
FROM quay.io/pypa/manylinux2014_${ARCH}
|
FROM quay.io/pypa/manylinux_2_28_${ARCH}
|
||||||
|
|
||||||
ARG ARCH=x86_64
|
ARG ARCH=x86_64
|
||||||
ARG DOCKER_USER=default_user
|
ARG DOCKER_USER=default_user
|
||||||
|
|||||||
0
ci/manylinux_nodejs/build.sh → ci/manylinux_node/build_lancedb.sh
Executable file → Normal file
0
ci/manylinux_nodejs/build.sh → ci/manylinux_node/build_lancedb.sh
Executable file → Normal file
@@ -6,7 +6,7 @@
|
|||||||
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
||||||
set -e
|
set -e
|
||||||
|
|
||||||
git clone -b OpenSSL_1_1_1u \
|
git clone -b OpenSSL_1_1_1v \
|
||||||
--single-branch \
|
--single-branch \
|
||||||
https://github.com/openssl/openssl.git
|
https://github.com/openssl/openssl.git
|
||||||
|
|
||||||
|
|||||||
@@ -8,7 +8,7 @@ install_node() {
|
|||||||
|
|
||||||
source "$HOME"/.bashrc
|
source "$HOME"/.bashrc
|
||||||
|
|
||||||
nvm install --no-progress 16
|
nvm install --no-progress 18
|
||||||
}
|
}
|
||||||
|
|
||||||
install_rust() {
|
install_rust() {
|
||||||
|
|||||||
@@ -1,31 +0,0 @@
|
|||||||
# Many linux dockerfile with Rust, Node, and Lance dependencies installed.
|
|
||||||
# This container allows building the node modules native libraries in an
|
|
||||||
# environment with a very old glibc, so that we are compatible with a wide
|
|
||||||
# range of linux distributions.
|
|
||||||
ARG ARCH=x86_64
|
|
||||||
|
|
||||||
FROM quay.io/pypa/manylinux2014_${ARCH}
|
|
||||||
|
|
||||||
ARG ARCH=x86_64
|
|
||||||
ARG DOCKER_USER=default_user
|
|
||||||
|
|
||||||
# Install static openssl
|
|
||||||
COPY install_openssl.sh install_openssl.sh
|
|
||||||
RUN ./install_openssl.sh ${ARCH} > /dev/null
|
|
||||||
|
|
||||||
# Protobuf is also installed as root.
|
|
||||||
COPY install_protobuf.sh install_protobuf.sh
|
|
||||||
RUN ./install_protobuf.sh ${ARCH}
|
|
||||||
|
|
||||||
ENV DOCKER_USER=${DOCKER_USER}
|
|
||||||
# Create a group and user
|
|
||||||
RUN echo ${ARCH} && adduser --user-group --create-home --uid ${DOCKER_USER} build_user
|
|
||||||
|
|
||||||
# We switch to the user to install Rust and Node, since those like to be
|
|
||||||
# installed at the user level.
|
|
||||||
USER ${DOCKER_USER}
|
|
||||||
|
|
||||||
COPY prepare_manylinux_node.sh prepare_manylinux_node.sh
|
|
||||||
RUN cp /prepare_manylinux_node.sh $HOME/ && \
|
|
||||||
cd $HOME && \
|
|
||||||
./prepare_manylinux_node.sh ${ARCH}
|
|
||||||
@@ -1,26 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
# Builds openssl from source so we can statically link to it
|
|
||||||
|
|
||||||
# this is to avoid the error we get with the system installation:
|
|
||||||
# /usr/bin/ld: <library>: version node not found for symbol SSLeay@@OPENSSL_1.0.1
|
|
||||||
# /usr/bin/ld: failed to set dynamic section sizes: Bad value
|
|
||||||
set -e
|
|
||||||
|
|
||||||
git clone -b OpenSSL_1_1_1u \
|
|
||||||
--single-branch \
|
|
||||||
https://github.com/openssl/openssl.git
|
|
||||||
|
|
||||||
pushd openssl
|
|
||||||
|
|
||||||
if [[ $1 == x86_64* ]]; then
|
|
||||||
ARCH=linux-x86_64
|
|
||||||
else
|
|
||||||
# gnu target
|
|
||||||
ARCH=linux-aarch64
|
|
||||||
fi
|
|
||||||
|
|
||||||
./Configure no-shared $ARCH
|
|
||||||
|
|
||||||
make
|
|
||||||
|
|
||||||
make install
|
|
||||||
@@ -1,15 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
# Installs protobuf compiler. Should be run as root.
|
|
||||||
set -e
|
|
||||||
|
|
||||||
if [[ $1 == x86_64* ]]; then
|
|
||||||
ARCH=x86_64
|
|
||||||
else
|
|
||||||
# gnu target
|
|
||||||
ARCH=aarch_64
|
|
||||||
fi
|
|
||||||
|
|
||||||
PB_REL=https://github.com/protocolbuffers/protobuf/releases
|
|
||||||
PB_VERSION=23.1
|
|
||||||
curl -LO $PB_REL/download/v$PB_VERSION/protoc-$PB_VERSION-linux-$ARCH.zip
|
|
||||||
unzip protoc-$PB_VERSION-linux-$ARCH.zip -d /usr/local
|
|
||||||
@@ -1,21 +0,0 @@
|
|||||||
#!/bin/bash
|
|
||||||
set -e
|
|
||||||
|
|
||||||
install_node() {
|
|
||||||
echo "Installing node..."
|
|
||||||
|
|
||||||
curl -o- https://raw.githubusercontent.com/nvm-sh/nvm/v0.34.0/install.sh | bash
|
|
||||||
|
|
||||||
source "$HOME"/.bashrc
|
|
||||||
|
|
||||||
nvm install --no-progress 16
|
|
||||||
}
|
|
||||||
|
|
||||||
install_rust() {
|
|
||||||
echo "Installing rust..."
|
|
||||||
curl https://sh.rustup.rs -sSf | bash -s -- -y
|
|
||||||
export PATH="$PATH:/root/.cargo/bin"
|
|
||||||
}
|
|
||||||
|
|
||||||
install_node
|
|
||||||
install_rust
|
|
||||||
@@ -141,10 +141,13 @@ nav:
|
|||||||
- Overview: examples/index.md
|
- Overview: examples/index.md
|
||||||
- 🐍 Python:
|
- 🐍 Python:
|
||||||
- Overview: examples/examples_python.md
|
- Overview: examples/examples_python.md
|
||||||
|
- Build From Scratch: examples/python_examples/build_from_scratch.md
|
||||||
|
- Multimodal: examples/python_examples/multimodal.md
|
||||||
|
- Rag: examples/python_examples/rag.md
|
||||||
|
- Miscellaneous:
|
||||||
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
||||||
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
||||||
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
||||||
- Example - Calculate CLIP Embeddings with Roboflow Inference: examples/image_embeddings_roboflow.md
|
|
||||||
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
||||||
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
||||||
- 👾 JavaScript:
|
- 👾 JavaScript:
|
||||||
@@ -221,14 +224,24 @@ nav:
|
|||||||
- PromptTools: integrations/prompttools.md
|
- PromptTools: integrations/prompttools.md
|
||||||
- Examples:
|
- Examples:
|
||||||
- examples/index.md
|
- examples/index.md
|
||||||
|
- 🐍 Python:
|
||||||
|
- Overview: examples/examples_python.md
|
||||||
|
- Build From Scratch: examples/python_examples/build_from_scratch.md
|
||||||
|
- Multimodal: examples/python_examples/multimodal.md
|
||||||
|
- Rag: examples/python_examples/rag.md
|
||||||
|
- Miscellaneous:
|
||||||
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
- YouTube Transcript Search: notebooks/youtube_transcript_search.ipynb
|
||||||
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
- Documentation QA Bot using LangChain: notebooks/code_qa_bot.ipynb
|
||||||
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
- Multimodal search using CLIP: notebooks/multimodal_search.ipynb
|
||||||
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
- Serverless QA Bot with S3 and Lambda: examples/serverless_lancedb_with_s3_and_lambda.md
|
||||||
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
- Serverless QA Bot with Modal: examples/serverless_qa_bot_with_modal_and_langchain.md
|
||||||
- YouTube Transcript Search (JS): examples/youtube_transcript_bot_with_nodejs.md
|
- 👾 JavaScript:
|
||||||
- Serverless Chatbot from any website: examples/serverless_website_chatbot.md
|
- Overview: examples/examples_js.md
|
||||||
|
- Serverless Website Chatbot: examples/serverless_website_chatbot.md
|
||||||
|
- YouTube Transcript Search: examples/youtube_transcript_bot_with_nodejs.md
|
||||||
- TransformersJS Embedding Search: examples/transformerjs_embedding_search_nodejs.md
|
- TransformersJS Embedding Search: examples/transformerjs_embedding_search_nodejs.md
|
||||||
|
- 🦀 Rust:
|
||||||
|
- Overview: examples/examples_rust.md
|
||||||
- API reference:
|
- API reference:
|
||||||
- Overview: api_reference.md
|
- Overview: api_reference.md
|
||||||
- Python: python/python.md
|
- Python: python/python.md
|
||||||
|
|||||||
1
docs/src/assets/colab.svg
Normal file
1
docs/src/assets/colab.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="117" height="20"><linearGradient id="b" x2="0" y2="100%"><stop offset="0" stop-color="#bbb" stop-opacity=".1"/><stop offset="1" stop-opacity=".1"/></linearGradient><clipPath id="a"><rect width="117" height="20" rx="3" fill="#fff"/></clipPath><g clip-path="url(#a)"><path fill="#555" d="M0 0h30v20H0z"/><path fill="#007ec6" d="M30 0h87v20H30z"/><path fill="url(#b)" d="M0 0h117v20H0z"/></g><g fill="#fff" text-anchor="middle" font-family="DejaVu Sans,Verdana,Geneva,sans-serif" font-size="110"><svg x="4px" y="0px" width="22px" height="20px" viewBox="-2 0 28 24" style="background-color: #fff;border-radius: 1px;"><path style="fill:#e8710a;" d="M1.977,16.77c-2.667-2.277-2.605-7.079,0-9.357C2.919,8.057,3.522,9.075,4.49,9.691c-1.152,1.6-1.146,3.201-0.004,4.803C3.522,15.111,2.918,16.126,1.977,16.77z"/><path style="fill:#f9ab00;" d="M12.257,17.114c-1.767-1.633-2.485-3.658-2.118-6.02c0.451-2.91,2.139-4.893,4.946-5.678c2.565-0.718,4.964-0.217,6.878,1.819c-0.884,0.743-1.707,1.547-2.434,2.446C18.488,8.827,17.319,8.435,16,8.856c-2.404,0.767-3.046,3.241-1.494,5.644c-0.241,0.275-0.493,0.541-0.721,0.826C13.295,15.939,12.511,16.3,12.257,17.114z"/><path style="fill:#e8710a;" d="M19.529,9.682c0.727-0.899,1.55-1.703,2.434-2.446c2.703,2.783,2.701,7.031-0.005,9.764c-2.648,2.674-6.936,2.725-9.701,0.115c0.254-0.814,1.038-1.175,1.528-1.788c0.228-0.285,0.48-0.552,0.721-0.826c1.053,0.916,2.254,1.268,3.6,0.83C20.502,14.551,21.151,11.927,19.529,9.682z"/><path style="fill:#f9ab00;" d="M4.49,9.691C3.522,9.075,2.919,8.057,1.977,7.413c2.209-2.398,5.721-2.942,8.476-1.355c0.555,0.32,0.719,0.606,0.285,1.128c-0.157,0.188-0.258,0.422-0.391,0.631c-0.299,0.47-0.509,1.067-0.929,1.371C8.933,9.539,8.523,8.847,8.021,8.746C6.673,8.475,5.509,8.787,4.49,9.691z"/><path style="fill:#f9ab00;" d="M1.977,16.77c0.941-0.644,1.545-1.659,2.509-2.277c1.373,1.152,2.85,1.433,4.45,0.499c0.332-0.194,0.503-0.088,0.673,0.19c0.386,0.635,0.753,1.285,1.181,1.89c0.34,0.48,0.222,0.715-0.253,1.006C7.84,19.73,4.205,19.188,1.977,16.77z"/></svg><text x="245" y="140" transform="scale(.1)" textLength="30"> </text><text x="725" y="150" fill="#010101" fill-opacity=".3" transform="scale(.1)" textLength="770">Open in Colab</text><text x="725" y="140" transform="scale(.1)" textLength="770">Open in Colab</text></g> </svg>
|
||||||
|
After Width: | Height: | Size: 2.3 KiB |
1
docs/src/assets/ghost.svg
Normal file
1
docs/src/assets/ghost.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="88.25" height="28" role="img" aria-label="GHOST"><title>GHOST</title><g shape-rendering="crispEdges"><rect width="88.25" height="28" fill="#000"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSIjZjdkZjFlIiByb2xlPSJpbWciIHZpZXdCb3g9IjAgMCAyNCAyNCIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIj48dGl0bGU+R2hvc3Q8L3RpdGxlPjxwYXRoIGQ9Ik0xMiAwQzUuMzczIDAgMCA1LjM3MyAwIDEyczUuMzczIDEyIDEyIDEyIDEyLTUuMzczIDEyLTEyUzE4LjYyNyAwIDEyIDB6bS4yNTYgMi4zMTNjMi40Ny4wMDUgNS4xMTYgMi4wMDggNS44OTggMi45NjJsLjI0NC4zYzEuNjQgMS45OTQgMy41NjkgNC4zNCAzLjU2OSA2Ljk2NiAwIDMuNzE5LTIuOTggNS44MDgtNi4xNTggNy41MDgtMS40MzMuNzY2LTIuOTggMS41MDgtNC43NDggMS41MDgtNC41NDMgMC04LjM2Ni0zLjU2OS04LjM2Ni04LjExMiAwLS43MDYuMTctMS40MjUuMzQyLTIuMTUuMTIyLS41MTUuMjQ0LTEuMDMzLjMwNy0xLjU0OS41NDgtNC41MzkgMi45NjctNi43OTUgOC40MjItNy40MDhhNC4yOSA0LjI5IDAgMDEuNDktLjAyNloiLz48L3N2Zz4="/><text transform="scale(.1)" x="541.25" y="175" textLength="442.5" fill="#fff" font-weight="bold">GHOST</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 1.2 KiB |
1
docs/src/assets/github.svg
Normal file
1
docs/src/assets/github.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="95.5" height="28" role="img" aria-label="GITHUB"><title>GITHUB</title><g shape-rendering="crispEdges"><rect width="95.5" height="28" fill="#121011"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSJ3aGl0ZSIgcm9sZT0iaW1nIiB2aWV3Qm94PSIwIDAgMjQgMjQiIHhtbG5zPSJodHRwOi8vd3d3LnczLm9yZy8yMDAwL3N2ZyI+PHRpdGxlPkdpdEh1YjwvdGl0bGU+PHBhdGggZD0iTTEyIC4yOTdjLTYuNjMgMC0xMiA1LjM3My0xMiAxMiAwIDUuMzAzIDMuNDM4IDkuOCA4LjIwNSAxMS4zODUuNi4xMTMuODItLjI1OC44Mi0uNTc3IDAtLjI4NS0uMDEtMS4wNC0uMDE1LTIuMDQtMy4zMzguNzI0LTQuMDQyLTEuNjEtNC4wNDItMS42MUM0LjQyMiAxOC4wNyAzLjYzMyAxNy43IDMuNjMzIDE3LjdjLTEuMDg3LS43NDQuMDg0LS43MjkuMDg0LS43MjkgMS4yMDUuMDg0IDEuODM4IDEuMjM2IDEuODM4IDEuMjM2IDEuMDcgMS44MzUgMi44MDkgMS4zMDUgMy40OTUuOTk4LjEwOC0uNzc2LjQxNy0xLjMwNS43Ni0xLjYwNS0yLjY2NS0uMy01LjQ2Ni0xLjMzMi01LjQ2Ni01LjkzIDAtMS4zMS40NjUtMi4zOCAxLjIzNS0zLjIyLS4xMzUtLjMwMy0uNTQtMS41MjMuMTA1LTMuMTc2IDAgMCAxLjAwNS0uMzIyIDMuMyAxLjIzLjk2LS4yNjcgMS45OC0uMzk5IDMtLjQwNSAxLjAyLjAwNiAyLjA0LjEzOCAzIC40MDUgMi4yOC0xLjU1MiAzLjI4NS0xLjIzIDMuMjg1LTEuMjMuNjQ1IDEuNjUzLjI0IDIuODczLjEyIDMuMTc2Ljc2NS44NCAxLjIzIDEuOTEgMS4yMyAzLjIyIDAgNC42MS0yLjgwNSA1LjYyNS01LjQ3NSA1LjkyLjQyLjM2LjgxIDEuMDk2LjgxIDIuMjIgMCAxLjYwNi0uMDE1IDIuODk2LS4wMTUgMy4yODYgMCAuMzE1LjIxLjY5LjgyNS41N0MyMC41NjUgMjIuMDkyIDI0IDE3LjU5MiAyNCAxMi4yOTdjMC02LjYyNy01LjM3My0xMi0xMi0xMiIvPjwvc3ZnPg=="/><text transform="scale(.1)" x="577.5" y="175" textLength="515" fill="#fff" font-weight="bold">GITHUB</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 1.7 KiB |
1
docs/src/assets/python.svg
Normal file
1
docs/src/assets/python.svg
Normal file
@@ -0,0 +1 @@
|
|||||||
|
<svg xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" width="97.5" height="28" role="img" aria-label="PYTHON"><title>PYTHON</title><g shape-rendering="crispEdges"><rect width="97.5" height="28" fill="#3670a0"/></g><g fill="#fff" text-anchor="middle" font-family="Verdana,Geneva,DejaVu Sans,sans-serif" text-rendering="geometricPrecision" font-size="100"><image x="9" y="7" width="14" height="14" xlink:href="data:image/svg+xml;base64,PHN2ZyBmaWxsPSIjZmZkZDU0IiByb2xlPSJpbWciIHZpZXdCb3g9IjAgMCAyNCAyNCIgeG1sbnM9Imh0dHA6Ly93d3cudzMub3JnLzIwMDAvc3ZnIj48dGl0bGU+UHl0aG9uPC90aXRsZT48cGF0aCBkPSJNMTQuMjUuMThsLjkuMi43My4yNi41OS4zLjQ1LjMyLjM0LjM0LjI1LjM0LjE2LjMzLjEuMy4wNC4yNi4wMi4yLS4wMS4xM1Y4LjVsLS4wNS42My0uMTMuNTUtLjIxLjQ2LS4yNi4zOC0uMy4zMS0uMzMuMjUtLjM1LjE5LS4zNS4xNC0uMzMuMS0uMy4wNy0uMjYuMDQtLjIxLjAySDguNzdsLS42OS4wNS0uNTkuMTQtLjUuMjItLjQxLjI3LS4zMy4zMi0uMjcuMzUtLjIuMzYtLjE1LjM3LS4xLjM1LS4wNy4zMi0uMDQuMjctLjAyLjIxdjMuMDZIMy4xN2wtLjIxLS4wMy0uMjgtLjA3LS4zMi0uMTItLjM1LS4xOC0uMzYtLjI2LS4zNi0uMzYtLjM1LS40Ni0uMzItLjU5LS4yOC0uNzMtLjIxLS44OC0uMTQtMS4wNS0uMDUtMS4yMy4wNi0xLjIyLjE2LTEuMDQuMjQtLjg3LjMyLS43MS4zNi0uNTcuNC0uNDQuNDItLjMzLjQyLS4yNC40LS4xNi4zNi0uMS4zMi0uMDUuMjQtLjAxaC4xNmwuMDYuMDFoOC4xNnYtLjgzSDYuMThsLS4wMS0yLjc1LS4wMi0uMzcuMDUtLjM0LjExLS4zMS4xNy0uMjguMjUtLjI2LjMxLS4yMy4zOC0uMi40NC0uMTguNTEtLjE1LjU4LS4xMi42NC0uMS43MS0uMDYuNzctLjA0Ljg0LS4wMiAxLjI3LjA1em0tNi4zIDEuOThsLS4yMy4zMy0uMDguNDEuMDguNDEuMjMuMzQuMzMuMjIuNDEuMDkuNDEtLjA5LjMzLS4yMi4yMy0uMzQuMDgtLjQxLS4wOC0uNDEtLjIzLS4zMy0uMzMtLjIyLS40MS0uMDktLjQxLjA5em0xMy4wOSAzLjk1bC4yOC4wNi4zMi4xMi4zNS4xOC4zNi4yNy4zNi4zNS4zNS40Ny4zMi41OS4yOC43My4yMS44OC4xNCAxLjA0LjA1IDEuMjMtLjA2IDEuMjMtLjE2IDEuMDQtLjI0Ljg2LS4zMi43MS0uMzYuNTctLjQuNDUtLjQyLjMzLS40Mi4yNC0uNC4xNi0uMzYuMDktLjMyLjA1LS4yNC4wMi0uMTYtLjAxaC04LjIydi44Mmg1Ljg0bC4wMSAyLjc2LjAyLjM2LS4wNS4zNC0uMTEuMzEtLjE3LjI5LS4yNS4yNS0uMzEuMjQtLjM4LjItLjQ0LjE3LS41MS4xNS0uNTguMTMtLjY0LjA5LS43MS4wNy0uNzcuMDQtLjg0LjAxLTEuMjctLjA0LTEuMDctLjE0LS45LS4yLS43My0uMjUtLjU5LS4zLS40NS0uMzMtLjM0LS4zNC0uMjUtLjM0LS4xNi0uMzMtLjEtLjMtLjA0LS4yNS0uMDItLjIuMDEtLjEzdi01LjM0bC4wNS0uNjQuMTMtLjU0LjIxLS40Ni4yNi0uMzguMy0uMzIuMzMtLjI0LjM1LS4yLjM1LS4xNC4zMy0uMS4zLS4wNi4yNi0uMDQuMjEtLjAyLjEzLS4wMWg1Ljg0bC42OS0uMDUuNTktLjE0LjUtLjIxLjQxLS4yOC4zMy0uMzIuMjctLjM1LjItLjM2LjE1LS4zNi4xLS4zNS4wNy0uMzIuMDQtLjI4LjAyLS4yMVY2LjA3aDIuMDlsLjE0LjAxem0tNi40NyAxNC4yNWwtLjIzLjMzLS4wOC40MS4wOC40MS4yMy4zMy4zMy4yMy40MS4wOC40MS0uMDguMzMtLjIzLjIzLS4zMy4wOC0uNDEtLjA4LS40MS0uMjMtLjMzLS4zMy0uMjMtLjQxLS4wOC0uNDEuMDh6Ii8+PC9zdmc+"/><text transform="scale(.1)" x="587.5" y="175" textLength="535" fill="#fff" font-weight="bold">PYTHON</text></g></svg>
|
||||||
|
After Width: | Height: | Size: 2.6 KiB |
@@ -15,12 +15,15 @@ There is another optional layer of abstraction available: `TextEmbeddingFunction
|
|||||||
|
|
||||||
Let's implement `SentenceTransformerEmbeddings` class. All you need to do is implement the `generate_embeddings()` and `ndims` function to handle the input types you expect and register the class in the global `EmbeddingFunctionRegistry`
|
Let's implement `SentenceTransformerEmbeddings` class. All you need to do is implement the `generate_embeddings()` and `ndims` function to handle the input types you expect and register the class in the global `EmbeddingFunctionRegistry`
|
||||||
|
|
||||||
```python
|
|
||||||
from lancedb.embeddings import register
|
|
||||||
from lancedb.util import attempt_import_or_raise
|
|
||||||
|
|
||||||
@register("sentence-transformers")
|
=== "Python"
|
||||||
class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
|
||||||
|
```python
|
||||||
|
from lancedb.embeddings import register
|
||||||
|
from lancedb.util import attempt_import_or_raise
|
||||||
|
|
||||||
|
@register("sentence-transformers")
|
||||||
|
class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
||||||
name: str = "all-MiniLM-L6-v2"
|
name: str = "all-MiniLM-L6-v2"
|
||||||
# set more default instance vars like device, etc.
|
# set more default instance vars like device, etc.
|
||||||
|
|
||||||
@@ -39,38 +42,59 @@ class SentenceTransformerEmbeddings(TextEmbeddingFunction):
|
|||||||
@cached(cache={})
|
@cached(cache={})
|
||||||
def _embedding_model(self):
|
def _embedding_model(self):
|
||||||
return sentence_transformers.SentenceTransformer(name)
|
return sentence_transformers.SentenceTransformer(name)
|
||||||
```
|
```
|
||||||
|
|
||||||
This is a stripped down version of our implementation of `SentenceTransformerEmbeddings` that removes certain optimizations and defaul settings.
|
=== "TypeScript"
|
||||||
|
|
||||||
|
```ts
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:imports"
|
||||||
|
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:embedding_impl"
|
||||||
|
```
|
||||||
|
|
||||||
|
|
||||||
|
This is a stripped down version of our implementation of `SentenceTransformerEmbeddings` that removes certain optimizations and default settings.
|
||||||
|
|
||||||
Now you can use this embedding function to create your table schema and that's it! you can then ingest data and run queries without manually vectorizing the inputs.
|
Now you can use this embedding function to create your table schema and that's it! you can then ingest data and run queries without manually vectorizing the inputs.
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
from lancedb.pydantic import LanceModel, Vector
|
|
||||||
|
|
||||||
registry = EmbeddingFunctionRegistry.get_instance()
|
```python
|
||||||
stransformer = registry.get("sentence-transformers").create()
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
|
||||||
class TextModelSchema(LanceModel):
|
registry = EmbeddingFunctionRegistry.get_instance()
|
||||||
|
stransformer = registry.get("sentence-transformers").create()
|
||||||
|
|
||||||
|
class TextModelSchema(LanceModel):
|
||||||
vector: Vector(stransformer.ndims) = stransformer.VectorField()
|
vector: Vector(stransformer.ndims) = stransformer.VectorField()
|
||||||
text: str = stransformer.SourceField()
|
text: str = stransformer.SourceField()
|
||||||
|
|
||||||
tbl = db.create_table("table", schema=TextModelSchema)
|
tbl = db.create_table("table", schema=TextModelSchema)
|
||||||
|
|
||||||
tbl.add(pd.DataFrame({"text": ["halo", "world"]}))
|
tbl.add(pd.DataFrame({"text": ["halo", "world"]}))
|
||||||
result = tbl.search("world").limit(5)
|
result = tbl.search("world").limit(5)
|
||||||
```
|
```
|
||||||
|
|
||||||
NOTE:
|
=== "TypeScript"
|
||||||
|
|
||||||
You can always implement the `EmbeddingFunction` interface directly if you want or need to, `TextEmbeddingFunction` just makes it much simpler and faster for you to do so, by setting up the boiler plat for text-specific use case
|
```ts
|
||||||
|
--8<--- "nodejs/examples/custom_embedding_function.ts:call_custom_function"
|
||||||
|
```
|
||||||
|
|
||||||
|
!!! note
|
||||||
|
|
||||||
|
You can always implement the `EmbeddingFunction` interface directly if you want or need to, `TextEmbeddingFunction` just makes it much simpler and faster for you to do so, by setting up the boiler plat for text-specific use case
|
||||||
|
|
||||||
## Multi-modal embedding function example
|
## Multi-modal embedding function example
|
||||||
You can also use the `EmbeddingFunction` interface to implement more complex workflows such as multi-modal embedding function support. LanceDB implements `OpenClipEmeddingFunction` class that suppports multi-modal seach. Here's the implementation that you can use as a reference to build your own multi-modal embedding functions.
|
You can also use the `EmbeddingFunction` interface to implement more complex workflows such as multi-modal embedding function support.
|
||||||
|
|
||||||
```python
|
=== "Python"
|
||||||
@register("open-clip")
|
|
||||||
class OpenClipEmbeddings(EmbeddingFunction):
|
LanceDB implements `OpenClipEmeddingFunction` class that suppports multi-modal seach. Here's the implementation that you can use as a reference to build your own multi-modal embedding functions.
|
||||||
|
|
||||||
|
```python
|
||||||
|
@register("open-clip")
|
||||||
|
class OpenClipEmbeddings(EmbeddingFunction):
|
||||||
name: str = "ViT-B-32"
|
name: str = "ViT-B-32"
|
||||||
pretrained: str = "laion2b_s34b_b79k"
|
pretrained: str = "laion2b_s34b_b79k"
|
||||||
device: str = "cpu"
|
device: str = "cpu"
|
||||||
@@ -209,4 +233,8 @@ class OpenClipEmbeddings(EmbeddingFunction):
|
|||||||
if self.normalize:
|
if self.normalize:
|
||||||
image_features /= image_features.norm(dim=-1, keepdim=True)
|
image_features /= image_features.norm(dim=-1, keepdim=True)
|
||||||
return image_features.cpu().numpy().squeeze()
|
return image_features.cpu().numpy().squeeze()
|
||||||
```
|
```
|
||||||
|
|
||||||
|
=== "TypeScript"
|
||||||
|
|
||||||
|
Coming Soon! See this [issue](https://github.com/lancedb/lancedb/issues/1482) to track the status!
|
||||||
|
|||||||
@@ -518,6 +518,82 @@ tbl.add(df)
|
|||||||
rs = tbl.search("hello").limit(1).to_pandas()
|
rs = tbl.search("hello").limit(1).to_pandas()
|
||||||
```
|
```
|
||||||
|
|
||||||
|
# IBM watsonx.ai Embeddings
|
||||||
|
|
||||||
|
Generate text embeddings using IBM's watsonx.ai platform.
|
||||||
|
|
||||||
|
## Supported Models
|
||||||
|
|
||||||
|
You can find a list of supported models at [IBM watsonx.ai Documentation](https://dataplatform.cloud.ibm.com/docs/content/wsj/analyze-data/fm-models-embed.html?context=wx). The currently supported model names are:
|
||||||
|
|
||||||
|
- `ibm/slate-125m-english-rtrvr`
|
||||||
|
- `ibm/slate-30m-english-rtrvr`
|
||||||
|
- `sentence-transformers/all-minilm-l12-v2`
|
||||||
|
- `intfloat/multilingual-e5-large`
|
||||||
|
|
||||||
|
## Parameters
|
||||||
|
|
||||||
|
The following parameters can be passed to the `create` method:
|
||||||
|
|
||||||
|
| Parameter | Type | Default Value | Description |
|
||||||
|
|------------|----------|----------------------------------|-----------------------------------------------------------|
|
||||||
|
| name | str | "ibm/slate-125m-english-rtrvr" | The model ID of the watsonx.ai model to use |
|
||||||
|
| api_key | str | None | Optional IBM Cloud API key (or set `WATSONX_API_KEY`) |
|
||||||
|
| project_id | str | None | Optional watsonx project ID (or set `WATSONX_PROJECT_ID`) |
|
||||||
|
| url | str | None | Optional custom URL for the watsonx.ai instance |
|
||||||
|
| params | dict | None | Optional additional parameters for the embedding model |
|
||||||
|
|
||||||
|
## Usage Example
|
||||||
|
|
||||||
|
First, the watsonx.ai library is an optional dependency, so must be installed seperately:
|
||||||
|
|
||||||
|
```
|
||||||
|
pip install ibm-watsonx-ai
|
||||||
|
```
|
||||||
|
|
||||||
|
Optionally set environment variables (if not passing credentials to `create` directly):
|
||||||
|
|
||||||
|
```sh
|
||||||
|
export WATSONX_API_KEY="YOUR_WATSONX_API_KEY"
|
||||||
|
export WATSONX_PROJECT_ID="YOUR_WATSONX_PROJECT_ID"
|
||||||
|
```
|
||||||
|
|
||||||
|
```python
|
||||||
|
import os
|
||||||
|
import lancedb
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.embeddings import EmbeddingFunctionRegistry
|
||||||
|
|
||||||
|
watsonx_embed = EmbeddingFunctionRegistry
|
||||||
|
.get_instance()
|
||||||
|
.get("watsonx")
|
||||||
|
.create(
|
||||||
|
name="ibm/slate-125m-english-rtrvr",
|
||||||
|
# Uncomment and set these if not using environment variables
|
||||||
|
# api_key="your_api_key_here",
|
||||||
|
# project_id="your_project_id_here",
|
||||||
|
# url="your_watsonx_url_here",
|
||||||
|
# params={...},
|
||||||
|
)
|
||||||
|
|
||||||
|
class TextModel(LanceModel):
|
||||||
|
text: str = watsonx_embed.SourceField()
|
||||||
|
vector: Vector(watsonx_embed.ndims()) = watsonx_embed.VectorField()
|
||||||
|
|
||||||
|
data = [
|
||||||
|
{"text": "hello world"},
|
||||||
|
{"text": "goodbye world"},
|
||||||
|
]
|
||||||
|
|
||||||
|
db = lancedb.connect("~/.lancedb")
|
||||||
|
tbl = db.create_table("watsonx_test", schema=TextModel, mode="overwrite")
|
||||||
|
|
||||||
|
tbl.add(data)
|
||||||
|
|
||||||
|
rs = tbl.search("hello").limit(1).to_pandas()
|
||||||
|
print(rs)
|
||||||
|
```
|
||||||
|
|
||||||
## Multi-modal embedding functions
|
## Multi-modal embedding functions
|
||||||
Multi-modal embedding functions allow you to query your table using both images and text.
|
Multi-modal embedding functions allow you to query your table using both images and text.
|
||||||
|
|
||||||
|
|||||||
@@ -10,7 +10,7 @@ LanceDB provides language APIs, allowing you to embed a database in your languag
|
|||||||
|
|
||||||
## Applications powered by LanceDB
|
## Applications powered by LanceDB
|
||||||
|
|
||||||
| Project Name | Description | Screenshot |
|
| Project Name | Description |
|
||||||
|-----------------------------------------------------|----------------------------------------------------------------------------------------------------------------------|-------------------------------------------|
|
| --- | --- |
|
||||||
| [YOLOExplorer](https://github.com/lancedb/yoloexplorer) | Iterate on your YOLO / CV datasets using SQL, Vector semantic search, and more within seconds | 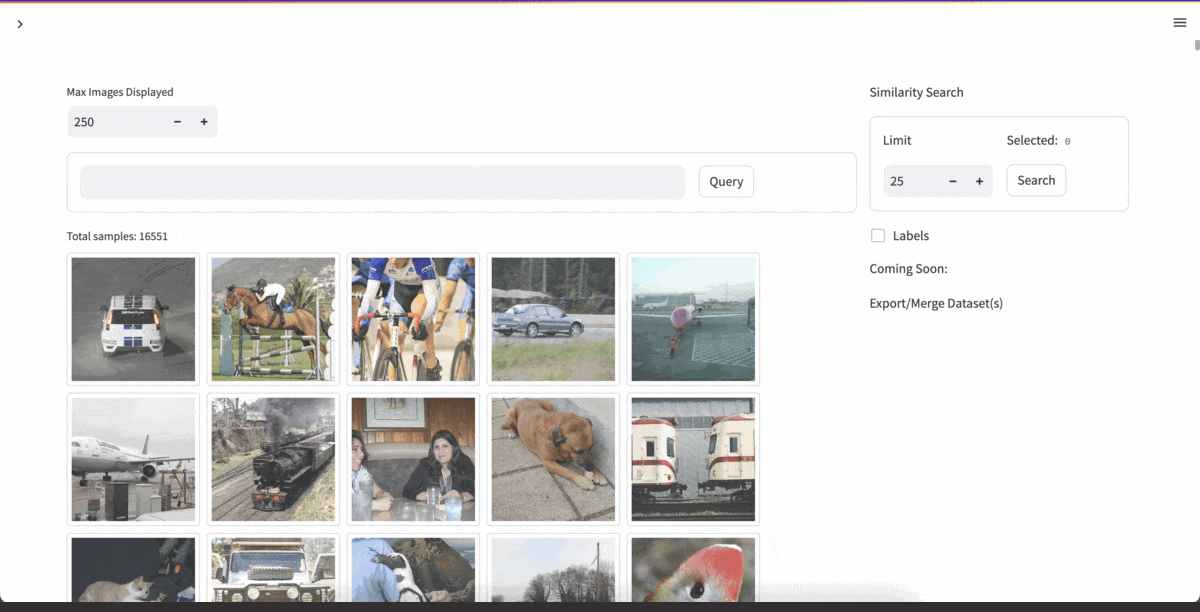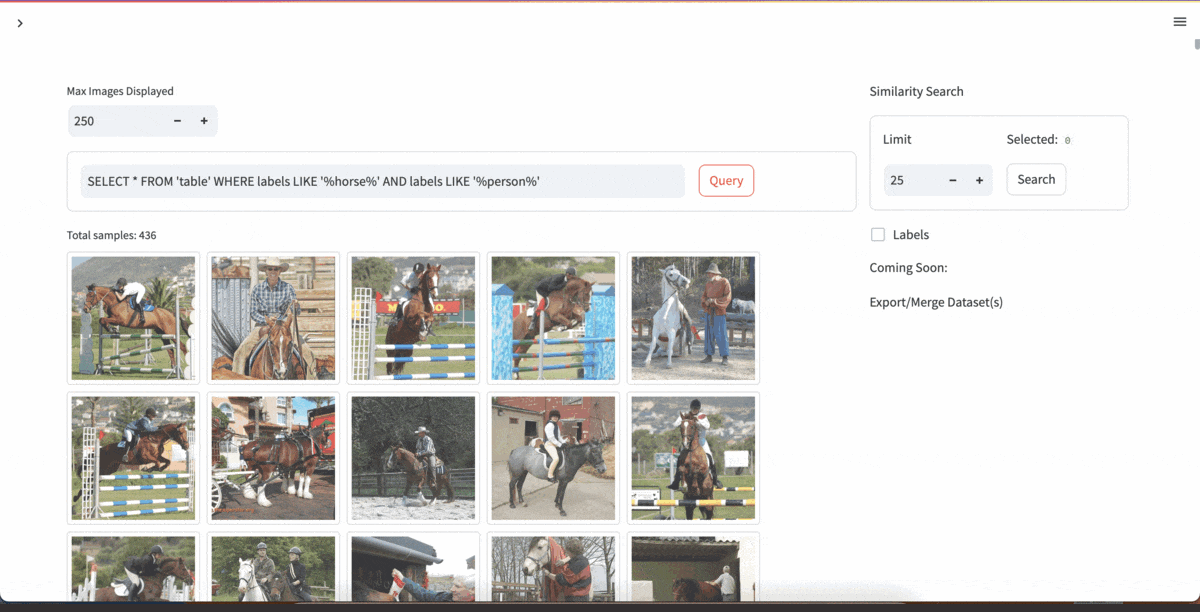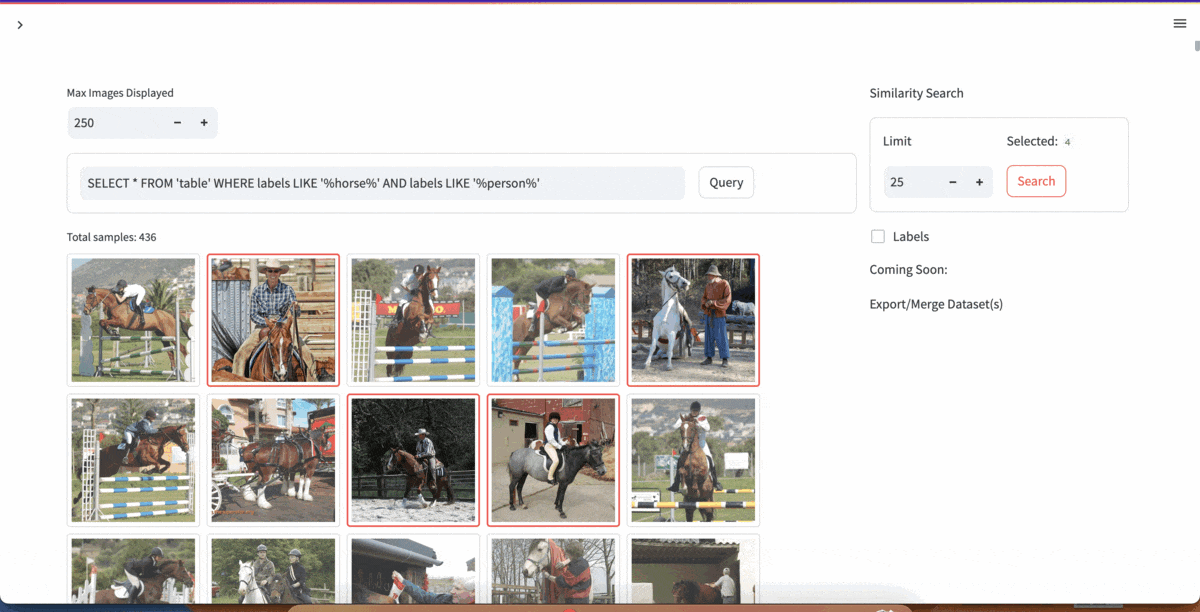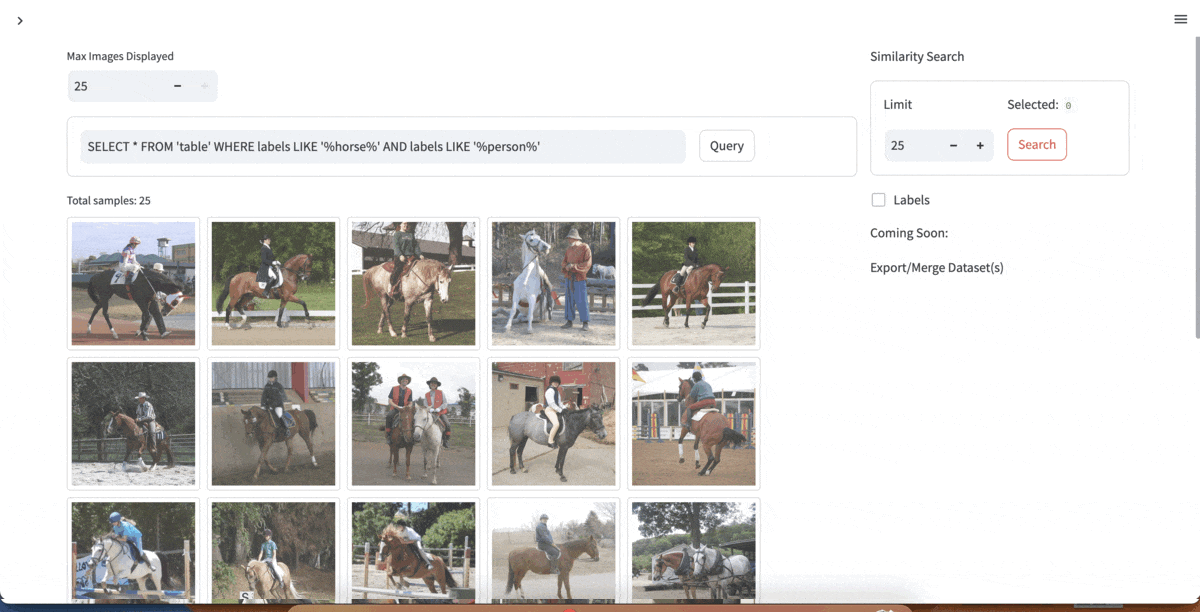 |
|
| **Ultralytics Explorer 🚀**<br>[](https://docs.ultralytics.com/datasets/explorer/)<br>[](https://colab.research.google.com/github/ultralytics/ultralytics/blob/main/docs/en/datasets/explorer/explorer.ipynb) | - 🔍 **Explore CV Datasets**: Semantic search, SQL queries, vector similarity, natural language.<br>- 🖥️ **GUI & Python API**: Seamless dataset interaction.<br>- ⚡ **Efficient & Scalable**: Leverages LanceDB for large datasets.<br>- 📊 **Detailed Analysis**: Easily analyze data patterns.<br>- 🌐 **Browser GUI Demo**: Create embeddings, search images, run queries. |
|
||||||
| [Website Chatbot (Deployable Vercel Template)](https://github.com/lancedb/lancedb-vercel-chatbot) | Create a chatbot from the sitemap of any website/docs of your choice. Built using vectorDB serverless native javascript package. |  |
|
| **Website Chatbot🤖**<br>[](https://github.com/lancedb/lancedb-vercel-chatbot)<br>[](https://vercel.com/new/clone?repository-url=https%3A%2F%2Fgithub.com%2Flancedb%2Flancedb-vercel-chatbot&env=OPENAI_API_KEY&envDescription=OpenAI%20API%20Key%20for%20chat%20completion.&project-name=lancedb-vercel-chatbot&repository-name=lancedb-vercel-chatbot&demo-title=LanceDB%20Chatbot%20Demo&demo-description=Demo%20website%20chatbot%20with%20LanceDB.&demo-url=https%3A%2F%2Flancedb.vercel.app&demo-image=https%3A%2F%2Fi.imgur.com%2FazVJtvr.png) | - 🌐 **Chatbot from Sitemap/Docs**: Create a chatbot using site or document context.<br>- 🚀 **Embed LanceDB in Next.js**: Lightweight, on-prem storage.<br>- 🧠 **AI-Powered Context Retrieval**: Efficiently access relevant data.<br>- 🔧 **Serverless & Native JS**: Seamless integration with Next.js.<br>- ⚡ **One-Click Deploy on Vercel**: Quick and easy setup.. |
|
||||||
|
|||||||
13
docs/src/examples/python_examples/build_from_scratch.md
Normal file
13
docs/src/examples/python_examples/build_from_scratch.md
Normal file
@@ -0,0 +1,13 @@
|
|||||||
|
# Build from Scratch with LanceDB 🚀
|
||||||
|
|
||||||
|
Start building your GenAI applications from the ground up using LanceDB's efficient vector-based document retrieval capabilities! 📄
|
||||||
|
|
||||||
|
#### Get Started in Minutes ⏱️
|
||||||
|
|
||||||
|
These examples provide a solid foundation for building your own GenAI applications using LanceDB. Jump from idea to proof of concept quickly with applied examples. Get started and see what you can create! 💻
|
||||||
|
|
||||||
|
| **Build From Scratch** | **Description** | **Links** |
|
||||||
|
|:-------------------------------------------|:-------------------------------------------------------------------------------------------------------------|:-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------|
|
||||||
|
| **Build RAG from Scratch🚀💻** | 📝 Create a **Retrieval-Augmented Generation** (RAG) model from scratch using LanceDB. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/RAG-from-Scratch)<br>[]() |
|
||||||
|
| **Local RAG from Scratch with Llama3🔥💡** | 🐫 Build a local RAG model using **Llama3** and **LanceDB** for fast and efficient text generation. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Local-RAG-from-Scratch)<br>[](https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Local-RAG-from-Scratch/rag.py) |
|
||||||
|
| **Multi-Head RAG from Scratch📚💻** | 🤯 Develop a **Multi-Head RAG model** from scratch, enabling generation of text based on multiple documents. | [](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Multi-Head-RAG-from-Scratch)<br>[](https://github.com/lancedb/vectordb-recipes/tree/main/tutorials/Multi-Head-RAG-from-Scratch) |
|
||||||
28
docs/src/examples/python_examples/multimodal.md
Normal file
28
docs/src/examples/python_examples/multimodal.md
Normal file
@@ -0,0 +1,28 @@
|
|||||||
|
# Multimodal Search with LanceDB 🔍💡
|
||||||
|
|
||||||
|
Experience the future of search with LanceDB's multimodal capabilities. Combine text and image queries to find the most relevant results in your corpus and unlock new possibilities! 🔓💡
|
||||||
|
|
||||||
|
#### Explore the Future of Search 🚀
|
||||||
|
|
||||||
|
Unlock the power of multimodal search with LanceDB, enabling efficient vector-based retrieval of text and image data! 📊💻
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
| **Multimodal** | **Description** | **Links** |
|
||||||
|
|:----------------|:-----------------|:-----------|
|
||||||
|
| **Multimodal CLIP: DiffusionDB 🌐💥** | Revolutionize search with Multimodal CLIP and DiffusionDB, combining text and image understanding for a new dimension of discovery! 🔓 | [][Clip_diffusionDB_github] <br>[][Clip_diffusionDB_colab] <br>[][Clip_diffusionDB_python] <br>[][Clip_diffusionDB_ghost] |
|
||||||
|
| **Multimodal CLIP: Youtube Videos 📹👀** | Search Youtube videos using Multimodal CLIP, finding relevant content with ease and accuracy! 🎯 | [][Clip_youtube_github] <br>[][Clip_youtube_colab] <br> [][Clip_youtube_python] <br>[][Clip_youtube_python] |
|
||||||
|
| **Multimodal Image + Text Search 📸🔍** | Discover relevant documents and images with a single query, using LanceDB's multimodal search capabilities to bridge the gap between text and visuals! 🌉 | [](https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_search) <br>[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_search/main.ipynb) <br> [](https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_search/main.py)<br> [](https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/) |
|
||||||
|
| **Cambrian-1: Vision-Centric Image Exploration 🔍👀** | Dive into vision-centric exploration of images with Cambrian-1, powered by LanceDB's multimodal search to uncover new insights! 🔎 | [](https://www.kaggle.com/code/prasantdixit/cambrian-1-vision-centric-exploration-of-images/)<br>[]() <br> []() <br> [](https://blog.lancedb.com/cambrian-1-vision-centric-exploration/) |
|
||||||
|
|
||||||
|
|
||||||
|
[Clip_diffusionDB_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb
|
||||||
|
[Clip_diffusionDB_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb/main.ipynb
|
||||||
|
[Clip_diffusionDB_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_clip_diffusiondb/main.py
|
||||||
|
[Clip_diffusionDB_ghost]: https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/
|
||||||
|
|
||||||
|
|
||||||
|
[Clip_youtube_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search
|
||||||
|
[Clip_youtube_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search/main.ipynb
|
||||||
|
[Clip_youtube_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/multimodal_video_search/main.py
|
||||||
|
[Clip_youtube_ghost]: https://blog.lancedb.com/multi-modal-ai-made-easy-with-lancedb-clip-5aaf8801c939/
|
||||||
85
docs/src/examples/python_examples/rag.md
Normal file
85
docs/src/examples/python_examples/rag.md
Normal file
@@ -0,0 +1,85 @@
|
|||||||
|
|
||||||
|
**🔍💡 RAG: Revolutionize Information Retrieval with LanceDB 🔓**
|
||||||
|
====================================================================
|
||||||
|
|
||||||
|
Unlock the full potential of Retrieval-Augmented Generation (RAG) with LanceDB, the ultimate solution for efficient vector-based information retrieval 📊. Input text queries and retrieve relevant documents with lightning-fast speed ⚡️ and accuracy ✅. Generate comprehensive answers by combining retrieved information, uncovering new insights 🔍 and connections.
|
||||||
|
|
||||||
|
### Experience the Future of Search 🔄
|
||||||
|
|
||||||
|
Experience the future of search with RAG, transforming information retrieval and answer generation. Apply RAG to various industries, streamlining processes 📈, saving time ⏰, and resources 💰. Stay ahead of the curve with innovative technology 🔝, powered by LanceDB. Discover the power of RAG with LanceDB and transform your industry with innovative solutions 💡.
|
||||||
|
|
||||||
|
|
||||||
|
| **RAG** | **Description** | **Links** |
|
||||||
|
|----------------------------------------------|------------------------------------------------------------------------------------------------------------------------------------------------------------------|----------------------------|
|
||||||
|
| **RAG with Matryoshka Embeddings and LlamaIndex** 🪆🔗 | Utilize **Matryoshka embeddings** and **LlamaIndex** to improve the efficiency and accuracy of your RAG models. 📈✨ | [][matryoshka_github] <br>[][matryoshka_colab] |
|
||||||
|
| **Improve RAG with Re-ranking** 📈🔄 | Enhance your RAG applications by implementing **re-ranking strategies** for more relevant document retrieval. 📚🔍 | [][rag_reranking_github] <br>[][rag_reranking_colab] <br>[][rag_reranking_ghost] |
|
||||||
|
| **Instruct-Multitask** 🧠🎯 | Integrate the **Instruct Embedding Model** with LanceDB to streamline your embedding API, reducing redundant code and overhead. 🌐📊 | [][instruct_multitask_github] <br>[][instruct_multitask_colab] <br>[][instruct_multitask_python] <br>[][instruct_multitask_ghost] |
|
||||||
|
| **Improve RAG with HyDE** 🌌🔍 | Use **Hypothetical Document Embeddings** for efficient, accurate, and unsupervised dense retrieval. 📄🔍 | [][hyde_github] <br>[][hyde_colab]<br>[][hyde_ghost] |
|
||||||
|
| **Improve RAG with LOTR** 🧙♂️📜 | Enhance RAG with **Lord of the Retriever (LOTR)** to address 'Lost in the Middle' challenges, especially in medical data. 🌟📜 | [][lotr_github] <br>[][lotr_colab] <br>[][lotr_ghost] |
|
||||||
|
| **Advanced RAG: Parent Document Retriever** 📑🔗 | Use **Parent Document & Bigger Chunk Retriever** to maintain context and relevance when generating related content. 🎵📄 | [][parent_doc_retriever_github] <br>[][parent_doc_retriever_colab] <br>[][parent_doc_retriever_ghost] |
|
||||||
|
| **Corrective RAG with Langgraph** 🔧📊 | Enhance RAG reliability with **Corrective RAG (CRAG)** by self-reflecting and fact-checking for accurate and trustworthy results. ✅🔍 |[][corrective_rag_github] <br>[][corrective_rag_colab] <br>[][corrective_rag_ghost] |
|
||||||
|
| **Contextual Compression with RAG** 🗜️🧠 | Apply **contextual compression techniques** to condense large documents while retaining essential information. 📄🗜️ | [][compression_rag_github] <br>[][compression_rag_colab] <br>[][compression_rag_ghost] |
|
||||||
|
| **Improve RAG with FLARE** 🔥| Enable users to ask questions directly to academic papers, focusing on ArXiv papers, with Forward-Looking Active REtrieval augmented generation.🚀🌟 | [][flare_github] <br>[][flare_colab] <br>[][flare_ghost] |
|
||||||
|
| **Query Expansion and Reranker** 🔍🔄 | Enhance RAG with query expansion using Large Language Models and advanced **reranking methods** like Cross Encoders, ColBERT v2, and FlashRank for improved document retrieval precision and recall 🔍📈 | [][query_github] <br>[][query_colab] |
|
||||||
|
| **RAG Fusion** ⚡🌐 | Revolutionize search with RAG Fusion, utilizing the **RRF algorithm** to rerank documents based on user queries, and leveraging LanceDB and OPENAI Embeddings for efficient information retrieval ⚡🌐 | [][fusion_github] <br>[][fusion_colab] |
|
||||||
|
| **Agentic RAG** 🤖📚 | Unlock autonomous information retrieval with **Agentic RAG**, a framework of **intelligent agents** that collaborate to synthesize, summarize, and compare data across sources, enabling proactive and informed decision-making 🤖📚 | [][agentic_github] <br>[][agentic_colab] |
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
[matryoshka_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/RAG-with_MatryoshkaEmbed-Llamaindex
|
||||||
|
[matryoshka_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/RAG-with_MatryoshkaEmbed-Llamaindex/RAG_with_MatryoshkaEmbedding_and_Llamaindex.ipynb
|
||||||
|
|
||||||
|
[rag_reranking_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/RAG_Reranking
|
||||||
|
[rag_reranking_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/RAG_Reranking/main.ipynb
|
||||||
|
[rag_reranking_ghost]: https://blog.lancedb.com/simplest-method-to-improve-rag-pipeline-re-ranking-cf6eaec6d544
|
||||||
|
|
||||||
|
|
||||||
|
[instruct_multitask_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask
|
||||||
|
[instruct_multitask_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask/main.ipynb
|
||||||
|
[instruct_multitask_python]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/instruct-multitask/main.py
|
||||||
|
[instruct_multitask_ghost]: https://blog.lancedb.com/multitask-embedding-with-lancedb-be18ec397543
|
||||||
|
|
||||||
|
[hyde_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE
|
||||||
|
[hyde_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE/main.ipynb
|
||||||
|
[hyde_ghost]: https://blog.lancedb.com/advanced-rag-precise-zero-shot-dense-retrieval-with-hyde-0946c54dfdcb
|
||||||
|
|
||||||
|
[lotr_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Advance_RAG_LOTR
|
||||||
|
[lotr_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance_RAG_LOTR/main.ipynb
|
||||||
|
[lotr_ghost]: https://blog.lancedb.com/better-rag-with-lotr-lord-of-retriever-23c8336b9a35
|
||||||
|
|
||||||
|
[parent_doc_retriever_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/parent_document_retriever
|
||||||
|
[parent_doc_retriever_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/parent_document_retriever/main.ipynb
|
||||||
|
[parent_doc_retriever_ghost]: https://blog.lancedb.com/modified-rag-parent-document-bigger-chunk-retriever-62b3d1e79bc6
|
||||||
|
|
||||||
|
[corrective_rag_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph
|
||||||
|
[corrective_rag_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph/CRAG_with_Langgraph.ipynb
|
||||||
|
[corrective_rag_ghost]: https://blog.lancedb.com/implementing-corrective-rag-in-the-easiest-way-2/
|
||||||
|
|
||||||
|
[compression_rag_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/Contextual-Compression-with-RAG
|
||||||
|
[compression_rag_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Contextual-Compression-with-RAG/main.ipynb
|
||||||
|
[compression_rag_ghost]: https://blog.lancedb.com/enhance-rag-integrate-contextual-compression-and-filtering-for-precision-a29d4a810301/
|
||||||
|
|
||||||
|
[flare_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR
|
||||||
|
[flare_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR/main.ipynb
|
||||||
|
[flare_ghost]: https://blog.lancedb.com/better-rag-with-active-retrieval-augmented-generation-flare-3b66646e2a9f/
|
||||||
|
|
||||||
|
[query_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/QueryExpansion&Reranker
|
||||||
|
[query_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/QueryExpansion&Reranker/main.ipynb
|
||||||
|
|
||||||
|
|
||||||
|
[fusion_github]: https://github.com/lancedb/vectordb-recipes/blob/main/examples/RAG_Fusion
|
||||||
|
[fusion_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/RAG_Fusion/main.ipynb
|
||||||
|
|
||||||
|
[agentic_github]: https://github.com/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG
|
||||||
|
[agentic_colab]: https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG/main.ipynb
|
||||||
|
|
||||||
|
|
||||||
@@ -5,4 +5,5 @@ pylance
|
|||||||
duckdb
|
duckdb
|
||||||
--extra-index-url https://download.pytorch.org/whl/cpu
|
--extra-index-url https://download.pytorch.org/whl/cpu
|
||||||
torch
|
torch
|
||||||
polars
|
polars>=0.19, <=1.3.0
|
||||||
|
|
||||||
|
|||||||
4
node/package-lock.json
generated
4
node/package-lock.json
generated
@@ -1,12 +1,12 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"lockfileVersion": 3,
|
"lockfileVersion": 3,
|
||||||
"requires": true,
|
"requires": true,
|
||||||
"packages": {
|
"packages": {
|
||||||
"": {
|
"": {
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"cpu": [
|
"cpu": [
|
||||||
"x64",
|
"x64",
|
||||||
"arm64"
|
"arm64"
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"description": " Serverless, low-latency vector database for AI applications",
|
"description": " Serverless, low-latency vector database for AI applications",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"types": "dist/index.d.ts",
|
"types": "dist/index.d.ts",
|
||||||
|
|||||||
@@ -20,7 +20,6 @@ napi = { version = "2.16.8", default-features = false, features = [
|
|||||||
"async",
|
"async",
|
||||||
] }
|
] }
|
||||||
napi-derive = "2.16.4"
|
napi-derive = "2.16.4"
|
||||||
|
|
||||||
# Prevent dynamic linking of lzma, which comes from datafusion
|
# Prevent dynamic linking of lzma, which comes from datafusion
|
||||||
lzma-sys = { version = "*", features = ["static"] }
|
lzma-sys = { version = "*", features = ["static"] }
|
||||||
|
|
||||||
|
|||||||
@@ -1,3 +1,4 @@
|
|||||||
|
import * as apiArrow from "apache-arrow";
|
||||||
// Copyright 2024 Lance Developers.
|
// Copyright 2024 Lance Developers.
|
||||||
//
|
//
|
||||||
// Licensed under the Apache License, Version 2.0 (the "License");
|
// Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
@@ -69,7 +70,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -82,7 +83,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
|
|
||||||
const schema = LanceSchema({
|
const schema = LanceSchema({
|
||||||
id: new arrow.Int32(),
|
id: new arrow.Int32(),
|
||||||
text: func.sourceField(new arrow.Utf8()),
|
text: func.sourceField(new arrow.Utf8() as apiArrow.DataType),
|
||||||
vector: func.vectorField(),
|
vector: func.vectorField(),
|
||||||
});
|
});
|
||||||
|
|
||||||
@@ -119,7 +120,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -144,7 +145,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
return 3;
|
return 3;
|
||||||
}
|
}
|
||||||
embeddingDataType() {
|
embeddingDataType() {
|
||||||
return new arrow.Float32();
|
return new arrow.Float32() as apiArrow.Float;
|
||||||
}
|
}
|
||||||
async computeSourceEmbeddings(data: string[]) {
|
async computeSourceEmbeddings(data: string[]) {
|
||||||
return data.map(() => [1, 2, 3]);
|
return data.map(() => [1, 2, 3]);
|
||||||
@@ -154,7 +155,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
|
|
||||||
const schema = LanceSchema({
|
const schema = LanceSchema({
|
||||||
id: new arrow.Int32(),
|
id: new arrow.Int32(),
|
||||||
text: func.sourceField(new arrow.Utf8()),
|
text: func.sourceField(new arrow.Utf8() as apiArrow.DataType),
|
||||||
vector: func.vectorField(),
|
vector: func.vectorField(),
|
||||||
});
|
});
|
||||||
const expectedMetadata = new Map<string, string>([
|
const expectedMetadata = new Map<string, string>([
|
||||||
|
|||||||
64
nodejs/examples/custom_embedding_function.ts
Normal file
64
nodejs/examples/custom_embedding_function.ts
Normal file
@@ -0,0 +1,64 @@
|
|||||||
|
// --8<-- [start:imports]
|
||||||
|
import * as lancedb from "@lancedb/lancedb";
|
||||||
|
import {
|
||||||
|
LanceSchema,
|
||||||
|
TextEmbeddingFunction,
|
||||||
|
getRegistry,
|
||||||
|
register,
|
||||||
|
} from "@lancedb/lancedb/embedding";
|
||||||
|
import { pipeline } from "@xenova/transformers";
|
||||||
|
// --8<-- [end:imports]
|
||||||
|
|
||||||
|
// --8<-- [start:embedding_impl]
|
||||||
|
@register("sentence-transformers")
|
||||||
|
class SentenceTransformersEmbeddings extends TextEmbeddingFunction {
|
||||||
|
name = "Xenova/all-miniLM-L6-v2";
|
||||||
|
#ndims!: number;
|
||||||
|
extractor: any;
|
||||||
|
|
||||||
|
async init() {
|
||||||
|
this.extractor = await pipeline("feature-extraction", this.name);
|
||||||
|
this.#ndims = await this.generateEmbeddings(["hello"]).then(
|
||||||
|
(e) => e[0].length,
|
||||||
|
);
|
||||||
|
}
|
||||||
|
|
||||||
|
ndims() {
|
||||||
|
return this.#ndims;
|
||||||
|
}
|
||||||
|
|
||||||
|
toJSON() {
|
||||||
|
return {
|
||||||
|
name: this.name,
|
||||||
|
};
|
||||||
|
}
|
||||||
|
async generateEmbeddings(texts: string[]) {
|
||||||
|
const output = await this.extractor(texts, {
|
||||||
|
pooling: "mean",
|
||||||
|
normalize: true,
|
||||||
|
});
|
||||||
|
return output.tolist();
|
||||||
|
}
|
||||||
|
}
|
||||||
|
// -8<-- [end:embedding_impl]
|
||||||
|
|
||||||
|
// --8<-- [start:call_custom_function]
|
||||||
|
const registry = getRegistry();
|
||||||
|
|
||||||
|
const sentenceTransformer = await registry
|
||||||
|
.get<SentenceTransformersEmbeddings>("sentence-transformers")!
|
||||||
|
.create();
|
||||||
|
|
||||||
|
const schema = LanceSchema({
|
||||||
|
vector: sentenceTransformer.vectorField(),
|
||||||
|
text: sentenceTransformer.sourceField(),
|
||||||
|
});
|
||||||
|
|
||||||
|
const db = await lancedb.connect("/tmp/db");
|
||||||
|
const table = await db.createEmptyTable("table", schema, { mode: "overwrite" });
|
||||||
|
|
||||||
|
await table.add([{ text: "hello" }, { text: "world" }]);
|
||||||
|
|
||||||
|
const results = await table.search("greeting").limit(1).toArray();
|
||||||
|
console.log(results[0].text);
|
||||||
|
// -8<-- [end:call_custom_function]
|
||||||
@@ -103,50 +103,11 @@ export type IntoVector =
|
|||||||
| number[]
|
| number[]
|
||||||
| Promise<Float32Array | Float64Array | number[]>;
|
| Promise<Float32Array | Float64Array | number[]>;
|
||||||
|
|
||||||
export type FloatLike =
|
|
||||||
| import("apache-arrow-13").Float
|
|
||||||
| import("apache-arrow-14").Float
|
|
||||||
| import("apache-arrow-15").Float
|
|
||||||
| import("apache-arrow-16").Float
|
|
||||||
| import("apache-arrow-17").Float;
|
|
||||||
export type DataTypeLike =
|
|
||||||
| import("apache-arrow-13").DataType
|
|
||||||
| import("apache-arrow-14").DataType
|
|
||||||
| import("apache-arrow-15").DataType
|
|
||||||
| import("apache-arrow-16").DataType
|
|
||||||
| import("apache-arrow-17").DataType;
|
|
||||||
|
|
||||||
export function isArrowTable(value: object): value is TableLike {
|
export function isArrowTable(value: object): value is TableLike {
|
||||||
if (value instanceof ArrowTable) return true;
|
if (value instanceof ArrowTable) return true;
|
||||||
return "schema" in value && "batches" in value;
|
return "schema" in value && "batches" in value;
|
||||||
}
|
}
|
||||||
|
|
||||||
export function isDataType(value: unknown): value is DataTypeLike {
|
|
||||||
return (
|
|
||||||
value instanceof DataType ||
|
|
||||||
DataType.isNull(value) ||
|
|
||||||
DataType.isInt(value) ||
|
|
||||||
DataType.isFloat(value) ||
|
|
||||||
DataType.isBinary(value) ||
|
|
||||||
DataType.isLargeBinary(value) ||
|
|
||||||
DataType.isUtf8(value) ||
|
|
||||||
DataType.isLargeUtf8(value) ||
|
|
||||||
DataType.isBool(value) ||
|
|
||||||
DataType.isDecimal(value) ||
|
|
||||||
DataType.isDate(value) ||
|
|
||||||
DataType.isTime(value) ||
|
|
||||||
DataType.isTimestamp(value) ||
|
|
||||||
DataType.isInterval(value) ||
|
|
||||||
DataType.isDuration(value) ||
|
|
||||||
DataType.isList(value) ||
|
|
||||||
DataType.isStruct(value) ||
|
|
||||||
DataType.isUnion(value) ||
|
|
||||||
DataType.isFixedSizeBinary(value) ||
|
|
||||||
DataType.isFixedSizeList(value) ||
|
|
||||||
DataType.isMap(value) ||
|
|
||||||
DataType.isDictionary(value)
|
|
||||||
);
|
|
||||||
}
|
|
||||||
export function isNull(value: unknown): value is Null {
|
export function isNull(value: unknown): value is Null {
|
||||||
return value instanceof Null || DataType.isNull(value);
|
return value instanceof Null || DataType.isNull(value);
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -44,10 +44,20 @@ export interface CreateTableOptions {
|
|||||||
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
||||||
*/
|
*/
|
||||||
storageOptions?: Record<string, string>;
|
storageOptions?: Record<string, string>;
|
||||||
|
/**
|
||||||
|
* The version of the data storage format to use.
|
||||||
|
*
|
||||||
|
* The default is `legacy`, which is Lance format v1.
|
||||||
|
* `stable` is the new format, which is Lance format v2.
|
||||||
|
*/
|
||||||
|
dataStorageVersion?: string;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* If true then data files will be written with the legacy format
|
* If true then data files will be written with the legacy format
|
||||||
*
|
*
|
||||||
* The default is true while the new format is in beta
|
* The default is true while the new format is in beta
|
||||||
|
*
|
||||||
|
* Deprecated.
|
||||||
*/
|
*/
|
||||||
useLegacyFormat?: boolean;
|
useLegacyFormat?: boolean;
|
||||||
schema?: SchemaLike;
|
schema?: SchemaLike;
|
||||||
@@ -247,12 +257,19 @@ export class LocalConnection extends Connection {
|
|||||||
throw new Error("data is required");
|
throw new Error("data is required");
|
||||||
}
|
}
|
||||||
const { buf, mode } = await Table.parseTableData(data, options);
|
const { buf, mode } = await Table.parseTableData(data, options);
|
||||||
|
let dataStorageVersion = "legacy";
|
||||||
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
|
dataStorageVersion = options.useLegacyFormat ? "legacy" : "stable";
|
||||||
|
}
|
||||||
|
|
||||||
const innerTable = await this.inner.createTable(
|
const innerTable = await this.inner.createTable(
|
||||||
nameOrOptions,
|
nameOrOptions,
|
||||||
buf,
|
buf,
|
||||||
mode,
|
mode,
|
||||||
cleanseStorageOptions(options?.storageOptions),
|
cleanseStorageOptions(options?.storageOptions),
|
||||||
options?.useLegacyFormat,
|
dataStorageVersion,
|
||||||
);
|
);
|
||||||
|
|
||||||
return new LocalTable(innerTable);
|
return new LocalTable(innerTable);
|
||||||
@@ -276,6 +293,13 @@ export class LocalConnection extends Connection {
|
|||||||
metadata = registry.getTableMetadata([embeddingFunction]);
|
metadata = registry.getTableMetadata([embeddingFunction]);
|
||||||
}
|
}
|
||||||
|
|
||||||
|
let dataStorageVersion = "legacy";
|
||||||
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
|
dataStorageVersion = options.useLegacyFormat ? "legacy" : "stable";
|
||||||
|
}
|
||||||
|
|
||||||
const table = makeEmptyTable(schema, metadata);
|
const table = makeEmptyTable(schema, metadata);
|
||||||
const buf = await fromTableToBuffer(table);
|
const buf = await fromTableToBuffer(table);
|
||||||
const innerTable = await this.inner.createEmptyTable(
|
const innerTable = await this.inner.createEmptyTable(
|
||||||
@@ -283,7 +307,7 @@ export class LocalConnection extends Connection {
|
|||||||
buf,
|
buf,
|
||||||
mode,
|
mode,
|
||||||
cleanseStorageOptions(options?.storageOptions),
|
cleanseStorageOptions(options?.storageOptions),
|
||||||
options?.useLegacyFormat,
|
dataStorageVersion,
|
||||||
);
|
);
|
||||||
return new LocalTable(innerTable);
|
return new LocalTable(innerTable);
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -15,13 +15,12 @@
|
|||||||
import "reflect-metadata";
|
import "reflect-metadata";
|
||||||
import {
|
import {
|
||||||
DataType,
|
DataType,
|
||||||
DataTypeLike,
|
|
||||||
Field,
|
Field,
|
||||||
FixedSizeList,
|
FixedSizeList,
|
||||||
|
Float,
|
||||||
Float32,
|
Float32,
|
||||||
FloatLike,
|
|
||||||
type IntoVector,
|
type IntoVector,
|
||||||
isDataType,
|
Utf8,
|
||||||
isFixedSizeList,
|
isFixedSizeList,
|
||||||
isFloat,
|
isFloat,
|
||||||
newVectorType,
|
newVectorType,
|
||||||
@@ -93,11 +92,12 @@ export abstract class EmbeddingFunction<
|
|||||||
* @see {@link lancedb.LanceSchema}
|
* @see {@link lancedb.LanceSchema}
|
||||||
*/
|
*/
|
||||||
sourceField(
|
sourceField(
|
||||||
optionsOrDatatype: Partial<FieldOptions> | DataTypeLike,
|
optionsOrDatatype: Partial<FieldOptions> | DataType,
|
||||||
): [DataTypeLike, Map<string, EmbeddingFunction>] {
|
): [DataType, Map<string, EmbeddingFunction>] {
|
||||||
let datatype = isDataType(optionsOrDatatype)
|
let datatype =
|
||||||
? optionsOrDatatype
|
"datatype" in optionsOrDatatype
|
||||||
: optionsOrDatatype?.datatype;
|
? optionsOrDatatype.datatype
|
||||||
|
: optionsOrDatatype;
|
||||||
if (!datatype) {
|
if (!datatype) {
|
||||||
throw new Error("Datatype is required");
|
throw new Error("Datatype is required");
|
||||||
}
|
}
|
||||||
@@ -123,15 +123,17 @@ export abstract class EmbeddingFunction<
|
|||||||
let dims: number | undefined = this.ndims();
|
let dims: number | undefined = this.ndims();
|
||||||
|
|
||||||
// `func.vectorField(new Float32())`
|
// `func.vectorField(new Float32())`
|
||||||
if (isDataType(optionsOrDatatype)) {
|
if (optionsOrDatatype === undefined) {
|
||||||
dtype = optionsOrDatatype;
|
dtype = new Float32();
|
||||||
|
} else if (!("datatype" in optionsOrDatatype)) {
|
||||||
|
dtype = sanitizeType(optionsOrDatatype);
|
||||||
} else {
|
} else {
|
||||||
// `func.vectorField({
|
// `func.vectorField({
|
||||||
// datatype: new Float32(),
|
// datatype: new Float32(),
|
||||||
// dims: 10
|
// dims: 10
|
||||||
// })`
|
// })`
|
||||||
dims = dims ?? optionsOrDatatype?.dims;
|
dims = dims ?? optionsOrDatatype?.dims;
|
||||||
dtype = optionsOrDatatype?.datatype;
|
dtype = sanitizeType(optionsOrDatatype?.datatype);
|
||||||
}
|
}
|
||||||
|
|
||||||
if (dtype !== undefined) {
|
if (dtype !== undefined) {
|
||||||
@@ -173,7 +175,7 @@ export abstract class EmbeddingFunction<
|
|||||||
}
|
}
|
||||||
|
|
||||||
/** The datatype of the embeddings */
|
/** The datatype of the embeddings */
|
||||||
abstract embeddingDataType(): FloatLike;
|
abstract embeddingDataType(): Float;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* Creates a vector representation for the given values.
|
* Creates a vector representation for the given values.
|
||||||
@@ -192,6 +194,38 @@ export abstract class EmbeddingFunction<
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* an abstract class for implementing embedding functions that take text as input
|
||||||
|
*/
|
||||||
|
export abstract class TextEmbeddingFunction<
|
||||||
|
M extends FunctionOptions = FunctionOptions,
|
||||||
|
> extends EmbeddingFunction<string, M> {
|
||||||
|
//** Generate the embeddings for the given texts */
|
||||||
|
abstract generateEmbeddings(
|
||||||
|
texts: string[],
|
||||||
|
// biome-ignore lint/suspicious/noExplicitAny: we don't know what the implementor will do
|
||||||
|
...args: any[]
|
||||||
|
): Promise<number[][] | Float32Array[] | Float64Array[]>;
|
||||||
|
|
||||||
|
async computeQueryEmbeddings(data: string): Promise<Awaited<IntoVector>> {
|
||||||
|
return this.generateEmbeddings([data]).then((data) => data[0]);
|
||||||
|
}
|
||||||
|
|
||||||
|
embeddingDataType(): Float {
|
||||||
|
return new Float32();
|
||||||
|
}
|
||||||
|
|
||||||
|
override sourceField(): [DataType, Map<string, EmbeddingFunction>] {
|
||||||
|
return super.sourceField(new Utf8());
|
||||||
|
}
|
||||||
|
|
||||||
|
computeSourceEmbeddings(
|
||||||
|
data: string[],
|
||||||
|
): Promise<number[][] | Float32Array[] | Float64Array[]> {
|
||||||
|
return this.generateEmbeddings(data);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
export interface FieldOptions<T extends DataType = DataType> {
|
export interface FieldOptions<T extends DataType = DataType> {
|
||||||
datatype: T;
|
datatype: T;
|
||||||
dims?: number;
|
dims?: number;
|
||||||
|
|||||||
@@ -13,12 +13,11 @@
|
|||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
import { Field, Schema } from "../arrow";
|
import { Field, Schema } from "../arrow";
|
||||||
import { isDataType } from "../arrow";
|
|
||||||
import { sanitizeType } from "../sanitize";
|
import { sanitizeType } from "../sanitize";
|
||||||
import { EmbeddingFunction } from "./embedding_function";
|
import { EmbeddingFunction } from "./embedding_function";
|
||||||
import { EmbeddingFunctionConfig, getRegistry } from "./registry";
|
import { EmbeddingFunctionConfig, getRegistry } from "./registry";
|
||||||
|
|
||||||
export { EmbeddingFunction } from "./embedding_function";
|
export { EmbeddingFunction, TextEmbeddingFunction } from "./embedding_function";
|
||||||
|
|
||||||
// We need to explicitly export '*' so that the `register` decorator actually registers the class.
|
// We need to explicitly export '*' so that the `register` decorator actually registers the class.
|
||||||
export * from "./openai";
|
export * from "./openai";
|
||||||
@@ -57,15 +56,15 @@ export function LanceSchema(
|
|||||||
Partial<EmbeddingFunctionConfig>
|
Partial<EmbeddingFunctionConfig>
|
||||||
>();
|
>();
|
||||||
Object.entries(fields).forEach(([key, value]) => {
|
Object.entries(fields).forEach(([key, value]) => {
|
||||||
if (isDataType(value)) {
|
if (Array.isArray(value)) {
|
||||||
arrowFields.push(new Field(key, sanitizeType(value), true));
|
|
||||||
} else {
|
|
||||||
const [dtype, metadata] = value as [
|
const [dtype, metadata] = value as [
|
||||||
object,
|
object,
|
||||||
Map<string, EmbeddingFunction>,
|
Map<string, EmbeddingFunction>,
|
||||||
];
|
];
|
||||||
arrowFields.push(new Field(key, sanitizeType(dtype), true));
|
arrowFields.push(new Field(key, sanitizeType(dtype), true));
|
||||||
parseEmbeddingFunctions(embeddingFunctions, key, metadata);
|
parseEmbeddingFunctions(embeddingFunctions, key, metadata);
|
||||||
|
} else {
|
||||||
|
arrowFields.push(new Field(key, sanitizeType(value), true));
|
||||||
}
|
}
|
||||||
});
|
});
|
||||||
const registry = getRegistry();
|
const registry = getRegistry();
|
||||||
|
|||||||
@@ -13,7 +13,7 @@
|
|||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
import type OpenAI from "openai";
|
import type OpenAI from "openai";
|
||||||
import { type EmbeddingCreateParams } from "openai/resources";
|
import type { EmbeddingCreateParams } from "openai/resources/index";
|
||||||
import { Float, Float32 } from "../arrow";
|
import { Float, Float32 } from "../arrow";
|
||||||
import { EmbeddingFunction } from "./embedding_function";
|
import { EmbeddingFunction } from "./embedding_function";
|
||||||
import { register } from "./registry";
|
import { register } from "./registry";
|
||||||
|
|||||||
@@ -59,7 +59,7 @@ export {
|
|||||||
|
|
||||||
export { Index, IndexOptions, IvfPqOptions } from "./indices";
|
export { Index, IndexOptions, IvfPqOptions } from "./indices";
|
||||||
|
|
||||||
export { Table, AddDataOptions, UpdateOptions } from "./table";
|
export { Table, AddDataOptions, UpdateOptions, OptimizeOptions } from "./table";
|
||||||
|
|
||||||
export * as embedding from "./embedding";
|
export * as embedding from "./embedding";
|
||||||
|
|
||||||
|
|||||||
@@ -340,8 +340,14 @@ export function sanitizeType(typeLike: unknown): DataType<any> {
|
|||||||
if (typeof typeLike !== "object" || typeLike === null) {
|
if (typeof typeLike !== "object" || typeLike === null) {
|
||||||
throw Error("Expected a Type but object was null/undefined");
|
throw Error("Expected a Type but object was null/undefined");
|
||||||
}
|
}
|
||||||
if (!("typeId" in typeLike) || !(typeof typeLike.typeId !== "function")) {
|
if (
|
||||||
throw Error("Expected a Type to have a typeId function");
|
!("typeId" in typeLike) ||
|
||||||
|
!(
|
||||||
|
typeof typeLike.typeId !== "function" ||
|
||||||
|
typeof typeLike.typeId !== "number"
|
||||||
|
)
|
||||||
|
) {
|
||||||
|
throw Error("Expected a Type to have a typeId property");
|
||||||
}
|
}
|
||||||
let typeId: Type;
|
let typeId: Type;
|
||||||
if (typeof typeLike.typeId === "function") {
|
if (typeof typeLike.typeId === "function") {
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-arm64",
|
"name": "@lancedb/lancedb-darwin-arm64",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.darwin-arm64.node",
|
"main": "lancedb.darwin-arm64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-x64",
|
"name": "@lancedb/lancedb-darwin-x64",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.darwin-x64.node",
|
"main": "lancedb.darwin-x64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.linux-arm64-gnu.node",
|
"main": "lancedb.linux-arm64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-x64-gnu",
|
"name": "@lancedb/lancedb-linux-x64-gnu",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.linux-x64-gnu.node",
|
"main": "lancedb.linux-x64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-win32-x64-msvc",
|
"name": "@lancedb/lancedb-win32-x64-msvc",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"os": ["win32"],
|
"os": ["win32"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.win32-x64-msvc.node",
|
"main": "lancedb.win32-x64-msvc.node",
|
||||||
|
|||||||
4
nodejs/package-lock.json
generated
4
nodejs/package-lock.json
generated
@@ -1,12 +1,12 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb",
|
"name": "@lancedb/lancedb",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"lockfileVersion": 3,
|
"lockfileVersion": 3,
|
||||||
"requires": true,
|
"requires": true,
|
||||||
"packages": {
|
"packages": {
|
||||||
"": {
|
"": {
|
||||||
"name": "@lancedb/lancedb",
|
"name": "@lancedb/lancedb",
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"cpu": [
|
"cpu": [
|
||||||
"x64",
|
"x64",
|
||||||
"arm64"
|
"arm64"
|
||||||
|
|||||||
@@ -10,7 +10,7 @@
|
|||||||
"vector database",
|
"vector database",
|
||||||
"ann"
|
"ann"
|
||||||
],
|
],
|
||||||
"version": "0.7.2",
|
"version": "0.8.0",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"exports": {
|
"exports": {
|
||||||
".": "./dist/index.js",
|
".": "./dist/index.js",
|
||||||
|
|||||||
@@ -13,13 +13,16 @@
|
|||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
use std::collections::HashMap;
|
use std::collections::HashMap;
|
||||||
|
use std::str::FromStr;
|
||||||
|
|
||||||
use napi::bindgen_prelude::*;
|
use napi::bindgen_prelude::*;
|
||||||
use napi_derive::*;
|
use napi_derive::*;
|
||||||
|
|
||||||
use crate::table::Table;
|
use crate::table::Table;
|
||||||
use crate::ConnectionOptions;
|
use crate::ConnectionOptions;
|
||||||
use lancedb::connection::{ConnectBuilder, Connection as LanceDBConnection, CreateTableMode};
|
use lancedb::connection::{
|
||||||
|
ConnectBuilder, Connection as LanceDBConnection, CreateTableMode, LanceFileVersion,
|
||||||
|
};
|
||||||
use lancedb::ipc::{ipc_file_to_batches, ipc_file_to_schema};
|
use lancedb::ipc::{ipc_file_to_batches, ipc_file_to_schema};
|
||||||
|
|
||||||
#[napi]
|
#[napi]
|
||||||
@@ -120,7 +123,7 @@ impl Connection {
|
|||||||
buf: Buffer,
|
buf: Buffer,
|
||||||
mode: String,
|
mode: String,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_options: Option<String>,
|
||||||
) -> napi::Result<Table> {
|
) -> napi::Result<Table> {
|
||||||
let batches = ipc_file_to_batches(buf.to_vec())
|
let batches = ipc_file_to_batches(buf.to_vec())
|
||||||
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
||||||
@@ -131,8 +134,11 @@ impl Connection {
|
|||||||
builder = builder.storage_option(key, value);
|
builder = builder.storage_option(key, value);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_option) = data_storage_options.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_option)
|
||||||
|
.map_err(|e| napi::Error::from_reason(format!("{}", e)))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
let tbl = builder
|
let tbl = builder
|
||||||
.execute()
|
.execute()
|
||||||
@@ -148,7 +154,7 @@ impl Connection {
|
|||||||
schema_buf: Buffer,
|
schema_buf: Buffer,
|
||||||
mode: String,
|
mode: String,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_options: Option<String>,
|
||||||
) -> napi::Result<Table> {
|
) -> napi::Result<Table> {
|
||||||
let schema = ipc_file_to_schema(schema_buf.to_vec()).map_err(|e| {
|
let schema = ipc_file_to_schema(schema_buf.to_vec()).map_err(|e| {
|
||||||
napi::Error::from_reason(format!("Failed to marshal schema from JS to Rust: {}", e))
|
napi::Error::from_reason(format!("Failed to marshal schema from JS to Rust: {}", e))
|
||||||
@@ -163,8 +169,11 @@ impl Connection {
|
|||||||
builder = builder.storage_option(key, value);
|
builder = builder.storage_option(key, value);
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_option) = data_storage_options.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_option)
|
||||||
|
.map_err(|e| napi::Error::from_reason(format!("{}", e)))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
let tbl = builder
|
let tbl = builder
|
||||||
.execute()
|
.execute()
|
||||||
|
|||||||
@@ -293,6 +293,7 @@ impl Table {
|
|||||||
.optimize(OptimizeAction::Prune {
|
.optimize(OptimizeAction::Prune {
|
||||||
older_than,
|
older_than,
|
||||||
delete_unverified: None,
|
delete_unverified: None,
|
||||||
|
error_if_tagged_old_versions: None,
|
||||||
})
|
})
|
||||||
.await
|
.await
|
||||||
.default_error()?
|
.default_error()?
|
||||||
|
|||||||
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.11.0"
|
current_version = "0.12.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb-python"
|
name = "lancedb-python"
|
||||||
version = "0.11.0"
|
version = "0.12.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
description = "Python bindings for LanceDB"
|
description = "Python bindings for LanceDB"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
|
|||||||
@@ -3,7 +3,7 @@ name = "lancedb"
|
|||||||
# version in Cargo.toml
|
# version in Cargo.toml
|
||||||
dependencies = [
|
dependencies = [
|
||||||
"deprecation",
|
"deprecation",
|
||||||
"pylance==0.15.0",
|
"pylance==0.16.0",
|
||||||
"ratelimiter~=1.0",
|
"ratelimiter~=1.0",
|
||||||
"requests>=2.31.0",
|
"requests>=2.31.0",
|
||||||
"retry>=0.9.2",
|
"retry>=0.9.2",
|
||||||
@@ -56,7 +56,7 @@ tests = [
|
|||||||
"pytest-asyncio",
|
"pytest-asyncio",
|
||||||
"duckdb",
|
"duckdb",
|
||||||
"pytz",
|
"pytz",
|
||||||
"polars>=0.19",
|
"polars>=0.19, <=1.3.0",
|
||||||
"tantivy",
|
"tantivy",
|
||||||
]
|
]
|
||||||
dev = ["ruff", "pre-commit"]
|
dev = ["ruff", "pre-commit"]
|
||||||
@@ -76,6 +76,7 @@ embeddings = [
|
|||||||
"awscli>=1.29.57",
|
"awscli>=1.29.57",
|
||||||
"botocore>=1.31.57",
|
"botocore>=1.31.57",
|
||||||
"ollama",
|
"ollama",
|
||||||
|
"ibm-watsonx-ai>=1.1.2",
|
||||||
]
|
]
|
||||||
azure = ["adlfs>=2024.2.0"]
|
azure = ["adlfs>=2024.2.0"]
|
||||||
|
|
||||||
|
|||||||
@@ -24,7 +24,7 @@ class Connection(object):
|
|||||||
mode: str,
|
mode: str,
|
||||||
data: pa.RecordBatchReader,
|
data: pa.RecordBatchReader,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
data_storage_version: Optional[str] = None,
|
||||||
) -> Table: ...
|
) -> Table: ...
|
||||||
async def create_empty_table(
|
async def create_empty_table(
|
||||||
self,
|
self,
|
||||||
@@ -32,7 +32,7 @@ class Connection(object):
|
|||||||
mode: str,
|
mode: str,
|
||||||
schema: pa.Schema,
|
schema: pa.Schema,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
data_storage_version: Optional[str] = None,
|
||||||
) -> Table: ...
|
) -> Table: ...
|
||||||
|
|
||||||
class Table:
|
class Table:
|
||||||
|
|||||||
@@ -560,6 +560,7 @@ class AsyncConnection(object):
|
|||||||
fill_value: Optional[float] = None,
|
fill_value: Optional[float] = None,
|
||||||
storage_options: Optional[Dict[str, str]] = None,
|
storage_options: Optional[Dict[str, str]] = None,
|
||||||
*,
|
*,
|
||||||
|
data_storage_version: Optional[str] = None,
|
||||||
use_legacy_format: Optional[bool] = None,
|
use_legacy_format: Optional[bool] = None,
|
||||||
) -> AsyncTable:
|
) -> AsyncTable:
|
||||||
"""Create an [AsyncTable][lancedb.table.AsyncTable] in the database.
|
"""Create an [AsyncTable][lancedb.table.AsyncTable] in the database.
|
||||||
@@ -603,9 +604,15 @@ class AsyncConnection(object):
|
|||||||
connection will be inherited by the table, but can be overridden here.
|
connection will be inherited by the table, but can be overridden here.
|
||||||
See available options at
|
See available options at
|
||||||
https://lancedb.github.io/lancedb/guides/storage/
|
https://lancedb.github.io/lancedb/guides/storage/
|
||||||
use_legacy_format: bool, optional, default True
|
data_storage_version: optional, str, default "legacy"
|
||||||
|
The version of the data storage format to use. Newer versions are more
|
||||||
|
efficient but require newer versions of lance to read. The default is
|
||||||
|
"legacy" which will use the legacy v1 version. See the user guide
|
||||||
|
for more details.
|
||||||
|
use_legacy_format: bool, optional, default True. (Deprecated)
|
||||||
If True, use the legacy format for the table. If False, use the new format.
|
If True, use the legacy format for the table. If False, use the new format.
|
||||||
The default is True while the new format is in beta.
|
The default is True while the new format is in beta.
|
||||||
|
This method is deprecated, use `data_storage_version` instead.
|
||||||
|
|
||||||
|
|
||||||
Returns
|
Returns
|
||||||
@@ -765,13 +772,18 @@ class AsyncConnection(object):
|
|||||||
if mode == "create" and exist_ok:
|
if mode == "create" and exist_ok:
|
||||||
mode = "exist_ok"
|
mode = "exist_ok"
|
||||||
|
|
||||||
|
if not data_storage_version:
|
||||||
|
data_storage_version = (
|
||||||
|
"legacy" if use_legacy_format is None or use_legacy_format else "stable"
|
||||||
|
)
|
||||||
|
|
||||||
if data is None:
|
if data is None:
|
||||||
new_table = await self._inner.create_empty_table(
|
new_table = await self._inner.create_empty_table(
|
||||||
name,
|
name,
|
||||||
mode,
|
mode,
|
||||||
schema,
|
schema,
|
||||||
storage_options=storage_options,
|
storage_options=storage_options,
|
||||||
use_legacy_format=use_legacy_format,
|
data_storage_version=data_storage_version,
|
||||||
)
|
)
|
||||||
else:
|
else:
|
||||||
data = data_to_reader(data, schema)
|
data = data_to_reader(data, schema)
|
||||||
@@ -780,7 +792,7 @@ class AsyncConnection(object):
|
|||||||
mode,
|
mode,
|
||||||
data,
|
data,
|
||||||
storage_options=storage_options,
|
storage_options=storage_options,
|
||||||
use_legacy_format=use_legacy_format,
|
data_storage_version=data_storage_version,
|
||||||
)
|
)
|
||||||
|
|
||||||
return AsyncTable(new_table)
|
return AsyncTable(new_table)
|
||||||
|
|||||||
@@ -26,3 +26,4 @@ from .transformers import TransformersEmbeddingFunction, ColbertEmbeddings
|
|||||||
from .imagebind import ImageBindEmbeddings
|
from .imagebind import ImageBindEmbeddings
|
||||||
from .utils import with_embeddings
|
from .utils import with_embeddings
|
||||||
from .jinaai import JinaEmbeddings
|
from .jinaai import JinaEmbeddings
|
||||||
|
from .watsonx import WatsonxEmbeddings
|
||||||
|
|||||||
111
python/python/lancedb/embeddings/watsonx.py
Normal file
111
python/python/lancedb/embeddings/watsonx.py
Normal file
@@ -0,0 +1,111 @@
|
|||||||
|
# Copyright (c) 2023. LanceDB Developers
|
||||||
|
#
|
||||||
|
# Licensed under the Apache License, Version 2.0 (the "License");
|
||||||
|
# you may not use this file except in compliance with the License.
|
||||||
|
# You may obtain a copy of the License at
|
||||||
|
# http://www.apache.org/licenses/LICENSE-2.0
|
||||||
|
#
|
||||||
|
# Unless required by applicable law or agreed to in writing, software
|
||||||
|
# distributed under the License is distributed on an "AS IS" BASIS,
|
||||||
|
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
||||||
|
# See the License for the specific language governing permissions and
|
||||||
|
# limitations under the License.
|
||||||
|
import os
|
||||||
|
from functools import cached_property
|
||||||
|
from typing import List, Optional, Dict, Union
|
||||||
|
|
||||||
|
from ..util import attempt_import_or_raise
|
||||||
|
from .base import TextEmbeddingFunction
|
||||||
|
from .registry import register
|
||||||
|
|
||||||
|
import numpy as np
|
||||||
|
|
||||||
|
DEFAULT_WATSONX_URL = "https://us-south.ml.cloud.ibm.com"
|
||||||
|
|
||||||
|
MODELS_DIMS = {
|
||||||
|
"ibm/slate-125m-english-rtrvr": 768,
|
||||||
|
"ibm/slate-30m-english-rtrvr": 384,
|
||||||
|
"sentence-transformers/all-minilm-l12-v2": 384,
|
||||||
|
"intfloat/multilingual-e5-large": 1024,
|
||||||
|
}
|
||||||
|
|
||||||
|
|
||||||
|
@register("watsonx")
|
||||||
|
class WatsonxEmbeddings(TextEmbeddingFunction):
|
||||||
|
"""
|
||||||
|
API Docs:
|
||||||
|
---------
|
||||||
|
https://cloud.ibm.com/apidocs/watsonx-ai#text-embeddings
|
||||||
|
|
||||||
|
Supported embedding models:
|
||||||
|
---------------------------
|
||||||
|
https://dataplatform.cloud.ibm.com/docs/content/wsj/analyze-data/fm-models-embed.html?context=wx
|
||||||
|
"""
|
||||||
|
|
||||||
|
name: str = "ibm/slate-125m-english-rtrvr"
|
||||||
|
api_key: Optional[str] = None
|
||||||
|
project_id: Optional[str] = None
|
||||||
|
url: Optional[str] = None
|
||||||
|
params: Optional[Dict] = None
|
||||||
|
|
||||||
|
@staticmethod
|
||||||
|
def model_names():
|
||||||
|
return [
|
||||||
|
"ibm/slate-125m-english-rtrvr",
|
||||||
|
"ibm/slate-30m-english-rtrvr",
|
||||||
|
"sentence-transformers/all-minilm-l12-v2",
|
||||||
|
"intfloat/multilingual-e5-large",
|
||||||
|
]
|
||||||
|
|
||||||
|
def ndims(self):
|
||||||
|
return self._ndims
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def _ndims(self):
|
||||||
|
if self.name not in MODELS_DIMS:
|
||||||
|
raise ValueError(f"Unknown model name {self.name}")
|
||||||
|
return MODELS_DIMS[self.name]
|
||||||
|
|
||||||
|
def generate_embeddings(
|
||||||
|
self,
|
||||||
|
texts: Union[List[str], np.ndarray],
|
||||||
|
*args,
|
||||||
|
**kwargs,
|
||||||
|
) -> List[List[float]]:
|
||||||
|
return self._watsonx_client.embed_documents(
|
||||||
|
texts=list(texts),
|
||||||
|
*args,
|
||||||
|
**kwargs,
|
||||||
|
)
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def _watsonx_client(self):
|
||||||
|
ibm_watsonx_ai = attempt_import_or_raise("ibm_watsonx_ai")
|
||||||
|
ibm_watsonx_ai_foundation_models = attempt_import_or_raise(
|
||||||
|
"ibm_watsonx_ai.foundation_models"
|
||||||
|
)
|
||||||
|
|
||||||
|
kwargs = {"model_id": self.name}
|
||||||
|
if self.params:
|
||||||
|
kwargs["params"] = self.params
|
||||||
|
if self.project_id:
|
||||||
|
kwargs["project_id"] = self.project_id
|
||||||
|
elif "WATSONX_PROJECT_ID" in os.environ:
|
||||||
|
kwargs["project_id"] = os.environ["WATSONX_PROJECT_ID"]
|
||||||
|
else:
|
||||||
|
raise ValueError("WATSONX_PROJECT_ID must be set or passed")
|
||||||
|
|
||||||
|
creds_kwargs = {}
|
||||||
|
if self.api_key:
|
||||||
|
creds_kwargs["api_key"] = self.api_key
|
||||||
|
elif "WATSONX_API_KEY" in os.environ:
|
||||||
|
creds_kwargs["api_key"] = os.environ["WATSONX_API_KEY"]
|
||||||
|
else:
|
||||||
|
raise ValueError("WATSONX_API_KEY must be set or passed")
|
||||||
|
if self.url:
|
||||||
|
creds_kwargs["url"] = self.url
|
||||||
|
else:
|
||||||
|
creds_kwargs["url"] = DEFAULT_WATSONX_URL
|
||||||
|
kwargs["credentials"] = ibm_watsonx_ai.Credentials(**creds_kwargs)
|
||||||
|
|
||||||
|
return ibm_watsonx_ai_foundation_models.Embeddings(**kwargs)
|
||||||
@@ -22,8 +22,9 @@ from lance import json_to_schema
|
|||||||
|
|
||||||
from lancedb.common import DATA, VEC, VECTOR_COLUMN_NAME
|
from lancedb.common import DATA, VEC, VECTOR_COLUMN_NAME
|
||||||
from lancedb.merge import LanceMergeInsertBuilder
|
from lancedb.merge import LanceMergeInsertBuilder
|
||||||
|
from lancedb.embeddings import EmbeddingFunctionRegistry
|
||||||
|
|
||||||
from ..query import LanceVectorQueryBuilder
|
from ..query import LanceVectorQueryBuilder, LanceQueryBuilder
|
||||||
from ..table import Query, Table, _sanitize_data
|
from ..table import Query, Table, _sanitize_data
|
||||||
from ..util import inf_vector_column_query, value_to_sql
|
from ..util import inf_vector_column_query, value_to_sql
|
||||||
from .arrow import to_ipc_binary
|
from .arrow import to_ipc_binary
|
||||||
@@ -58,6 +59,21 @@ class RemoteTable(Table):
|
|||||||
resp = self._conn._client.post(f"/v1/table/{self._name}/describe/")
|
resp = self._conn._client.post(f"/v1/table/{self._name}/describe/")
|
||||||
return resp["version"]
|
return resp["version"]
|
||||||
|
|
||||||
|
@cached_property
|
||||||
|
def embedding_functions(self) -> dict:
|
||||||
|
"""
|
||||||
|
Get the embedding functions for the table
|
||||||
|
|
||||||
|
Returns
|
||||||
|
-------
|
||||||
|
funcs: dict
|
||||||
|
A mapping of the vector column to the embedding function
|
||||||
|
or empty dict if not configured.
|
||||||
|
"""
|
||||||
|
return EmbeddingFunctionRegistry.get_instance().parse_functions(
|
||||||
|
self.schema.metadata
|
||||||
|
)
|
||||||
|
|
||||||
def to_arrow(self) -> pa.Table:
|
def to_arrow(self) -> pa.Table:
|
||||||
"""to_arrow() is not yet supported on LanceDB cloud."""
|
"""to_arrow() is not yet supported on LanceDB cloud."""
|
||||||
raise NotImplementedError("to_arrow() is not yet supported on LanceDB cloud.")
|
raise NotImplementedError("to_arrow() is not yet supported on LanceDB cloud.")
|
||||||
@@ -213,7 +229,7 @@ class RemoteTable(Table):
|
|||||||
data, _ = _sanitize_data(
|
data, _ = _sanitize_data(
|
||||||
data,
|
data,
|
||||||
self.schema,
|
self.schema,
|
||||||
metadata=None,
|
metadata=self.schema.metadata,
|
||||||
on_bad_vectors=on_bad_vectors,
|
on_bad_vectors=on_bad_vectors,
|
||||||
fill_value=fill_value,
|
fill_value=fill_value,
|
||||||
)
|
)
|
||||||
@@ -293,6 +309,7 @@ class RemoteTable(Table):
|
|||||||
"""
|
"""
|
||||||
if vector_column_name is None:
|
if vector_column_name is None:
|
||||||
vector_column_name = inf_vector_column_query(self.schema)
|
vector_column_name = inf_vector_column_query(self.schema)
|
||||||
|
query = LanceQueryBuilder._query_to_vector(self, query, vector_column_name)
|
||||||
return LanceVectorQueryBuilder(self, query, vector_column_name)
|
return LanceVectorQueryBuilder(self, query, vector_column_name)
|
||||||
|
|
||||||
def _execute_query(
|
def _execute_query(
|
||||||
@@ -336,7 +353,7 @@ class RemoteTable(Table):
|
|||||||
|
|
||||||
See [`Table.merge_insert`][lancedb.table.Table.merge_insert] for more details.
|
See [`Table.merge_insert`][lancedb.table.Table.merge_insert] for more details.
|
||||||
"""
|
"""
|
||||||
super().merge_insert(on)
|
return super().merge_insert(on)
|
||||||
|
|
||||||
def _do_merge(
|
def _do_merge(
|
||||||
self,
|
self,
|
||||||
|
|||||||
@@ -1,9 +1,13 @@
|
|||||||
from abc import ABC, abstractmethod
|
from abc import ABC, abstractmethod
|
||||||
from packaging.version import Version
|
from packaging.version import Version
|
||||||
|
from typing import Union, List, TYPE_CHECKING
|
||||||
|
|
||||||
import numpy as np
|
import numpy as np
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
|
|
||||||
|
if TYPE_CHECKING:
|
||||||
|
from ..table import LanceVectorQueryBuilder
|
||||||
|
|
||||||
ARROW_VERSION = Version(pa.__version__)
|
ARROW_VERSION = Version(pa.__version__)
|
||||||
|
|
||||||
|
|
||||||
@@ -130,12 +134,94 @@ class Reranker(ABC):
|
|||||||
combined = pa.concat_tables(
|
combined = pa.concat_tables(
|
||||||
[vector_results, fts_results], **self._concat_tables_args
|
[vector_results, fts_results], **self._concat_tables_args
|
||||||
)
|
)
|
||||||
row_id = combined.column("_rowid")
|
|
||||||
|
|
||||||
# deduplicate
|
# deduplicate
|
||||||
mask = np.full((combined.shape[0]), False)
|
combined = self._deduplicate(combined)
|
||||||
_, mask_indices = np.unique(np.array(row_id), return_index=True)
|
|
||||||
mask[mask_indices] = True
|
|
||||||
combined = combined.filter(mask=mask)
|
|
||||||
|
|
||||||
return combined
|
return combined
|
||||||
|
|
||||||
|
def rerank_multivector(
|
||||||
|
self,
|
||||||
|
vector_results: Union[List[pa.Table], List["LanceVectorQueryBuilder"]],
|
||||||
|
query: Union[str, None], # Some rerankers might not need the query
|
||||||
|
deduplicate: bool = False,
|
||||||
|
):
|
||||||
|
"""
|
||||||
|
This is a rerank function that receives the results from multiple
|
||||||
|
vector searches. For example, this can be used to combine the
|
||||||
|
results of two vector searches with different embeddings.
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
vector_results : List[pa.Table] or List[LanceVectorQueryBuilder]
|
||||||
|
The results from the vector search. Either accepts the query builder
|
||||||
|
if the results haven't been executed yet or the results in arrow format.
|
||||||
|
query : str or None,
|
||||||
|
The input query. Some rerankers might not need the query to rerank.
|
||||||
|
In that case, it can be set to None explicitly. This is inteded to
|
||||||
|
be handled by the reranker implementations.
|
||||||
|
deduplicate : bool, optional
|
||||||
|
Whether to deduplicate the results based on the `_rowid` column,
|
||||||
|
by default False. Requires `_rowid` to be present in the results.
|
||||||
|
|
||||||
|
Returns
|
||||||
|
-------
|
||||||
|
pa.Table
|
||||||
|
The reranked results
|
||||||
|
"""
|
||||||
|
vector_results = (
|
||||||
|
[vector_results] if not isinstance(vector_results, list) else vector_results
|
||||||
|
)
|
||||||
|
|
||||||
|
# Make sure all elements are of the same type
|
||||||
|
if not all(isinstance(v, type(vector_results[0])) for v in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"All elements in vector_results should be of the same type"
|
||||||
|
)
|
||||||
|
|
||||||
|
# avoids circular import
|
||||||
|
if type(vector_results[0]).__name__ == "LanceVectorQueryBuilder":
|
||||||
|
vector_results = [result.to_arrow() for result in vector_results]
|
||||||
|
elif not isinstance(vector_results[0], pa.Table):
|
||||||
|
raise ValueError(
|
||||||
|
"vector_results should be a list of pa.Table or LanceVectorQueryBuilder"
|
||||||
|
)
|
||||||
|
|
||||||
|
combined = pa.concat_tables(vector_results, **self._concat_tables_args)
|
||||||
|
|
||||||
|
reranked = self.rerank_vector(query, combined)
|
||||||
|
|
||||||
|
# TODO: Allow custom deduplicators here.
|
||||||
|
# currently, this'll just keep the first instance.
|
||||||
|
if deduplicate:
|
||||||
|
if "_rowid" not in combined.column_names:
|
||||||
|
raise ValueError(
|
||||||
|
"'_rowid' is required for deduplication. \
|
||||||
|
add _rowid to search results like this: \
|
||||||
|
`search().with_row_id(True)`"
|
||||||
|
)
|
||||||
|
reranked = self._deduplicate(reranked)
|
||||||
|
|
||||||
|
return reranked
|
||||||
|
|
||||||
|
def _deduplicate(self, table: pa.Table):
|
||||||
|
"""
|
||||||
|
Deduplicate the table based on the `_rowid` column.
|
||||||
|
"""
|
||||||
|
row_id = table.column("_rowid")
|
||||||
|
|
||||||
|
# deduplicate
|
||||||
|
mask = np.full((table.shape[0]), False)
|
||||||
|
_, mask_indices = np.unique(np.array(row_id), return_index=True)
|
||||||
|
mask[mask_indices] = True
|
||||||
|
deduped_table = table.filter(mask=mask)
|
||||||
|
|
||||||
|
return deduped_table
|
||||||
|
|
||||||
|
def _keep_relevance_score(self, combined_results: pa.Table):
|
||||||
|
if self.score == "relevance":
|
||||||
|
if "score" in combined_results.column_names:
|
||||||
|
combined_results = combined_results.drop_columns(["score"])
|
||||||
|
if "_distance" in combined_results.column_names:
|
||||||
|
combined_results = combined_results.drop_columns(["_distance"])
|
||||||
|
return combined_results
|
||||||
|
|||||||
@@ -88,7 +88,7 @@ class CohereReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for cohere reranker"
|
"return_score='all' not implemented for cohere reranker"
|
||||||
|
|||||||
@@ -73,7 +73,7 @@ class ColbertReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"OpenAI Reranker does not support score='all' yet"
|
"OpenAI Reranker does not support score='all' yet"
|
||||||
|
|||||||
@@ -66,7 +66,7 @@ class CrossEncoderReranker(Reranker):
|
|||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
# sort the results by _score
|
# sort the results by _score
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for CrossEncoderReranker"
|
"return_score='all' not implemented for CrossEncoderReranker"
|
||||||
|
|||||||
@@ -92,7 +92,7 @@ class JinaReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"return_score='all' not implemented for JinaReranker"
|
"return_score='all' not implemented for JinaReranker"
|
||||||
|
|||||||
@@ -103,7 +103,7 @@ class LinearCombinationReranker(Reranker):
|
|||||||
[("_relevance_score", "descending")]
|
[("_relevance_score", "descending")]
|
||||||
)
|
)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
tbl = tbl.drop_columns(["score", "_distance"])
|
tbl = self._keep_relevance_score(tbl)
|
||||||
return tbl
|
return tbl
|
||||||
|
|
||||||
def _combine_score(self, score1, score2):
|
def _combine_score(self, score1, score2):
|
||||||
|
|||||||
@@ -84,7 +84,7 @@ class OpenaiReranker(Reranker):
|
|||||||
combined_results = self.merge_results(vector_results, fts_results)
|
combined_results = self.merge_results(vector_results, fts_results)
|
||||||
combined_results = self._rerank(combined_results, query)
|
combined_results = self._rerank(combined_results, query)
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
elif self.score == "all":
|
elif self.score == "all":
|
||||||
raise NotImplementedError(
|
raise NotImplementedError(
|
||||||
"OpenAI Reranker does not support score='all' yet"
|
"OpenAI Reranker does not support score='all' yet"
|
||||||
|
|||||||
@@ -1,8 +1,12 @@
|
|||||||
|
from typing import Union, List, TYPE_CHECKING
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
|
|
||||||
from collections import defaultdict
|
from collections import defaultdict
|
||||||
from .base import Reranker
|
from .base import Reranker
|
||||||
|
|
||||||
|
if TYPE_CHECKING:
|
||||||
|
from ..table import LanceVectorQueryBuilder
|
||||||
|
|
||||||
|
|
||||||
class RRFReranker(Reranker):
|
class RRFReranker(Reranker):
|
||||||
"""
|
"""
|
||||||
@@ -55,6 +59,46 @@ class RRFReranker(Reranker):
|
|||||||
)
|
)
|
||||||
|
|
||||||
if self.score == "relevance":
|
if self.score == "relevance":
|
||||||
combined_results = combined_results.drop_columns(["score", "_distance"])
|
combined_results = self._keep_relevance_score(combined_results)
|
||||||
|
|
||||||
return combined_results
|
return combined_results
|
||||||
|
|
||||||
|
def rerank_multivector(
|
||||||
|
self,
|
||||||
|
vector_results: Union[List[pa.Table], List["LanceVectorQueryBuilder"]],
|
||||||
|
query: str = None,
|
||||||
|
deduplicate: bool = True, # noqa: F821 # TODO: automatically deduplicates
|
||||||
|
):
|
||||||
|
"""
|
||||||
|
Overridden method to rerank the results from multiple vector searches.
|
||||||
|
This leverages the RRF hybrid reranking algorithm to combine the
|
||||||
|
results from multiple vector searches as it doesn't support reranking
|
||||||
|
vector results individually.
|
||||||
|
"""
|
||||||
|
# Make sure all elements are of the same type
|
||||||
|
if not all(isinstance(v, type(vector_results[0])) for v in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"All elements in vector_results should be of the same type"
|
||||||
|
)
|
||||||
|
|
||||||
|
# avoid circular import
|
||||||
|
if type(vector_results[0]).__name__ == "LanceVectorQueryBuilder":
|
||||||
|
vector_results = [result.to_arrow() for result in vector_results]
|
||||||
|
elif not isinstance(vector_results[0], pa.Table):
|
||||||
|
raise ValueError(
|
||||||
|
"vector_results should be a list of pa.Table or LanceVectorQueryBuilder"
|
||||||
|
)
|
||||||
|
|
||||||
|
# _rowid is required for RRF reranking
|
||||||
|
if not all("_rowid" in result.column_names for result in vector_results):
|
||||||
|
raise ValueError(
|
||||||
|
"'_rowid' is required for deduplication. \
|
||||||
|
add _rowid to search results like this: \
|
||||||
|
`search().with_row_id(True)`"
|
||||||
|
)
|
||||||
|
|
||||||
|
combined = pa.concat_tables(vector_results, **self._concat_tables_args)
|
||||||
|
empty_table = pa.Table.from_arrays([], names=[])
|
||||||
|
reranked = self.rerank_hybrid(query, combined, empty_table)
|
||||||
|
|
||||||
|
return reranked
|
||||||
|
|||||||
@@ -417,3 +417,28 @@ def test_openai_embedding(tmp_path):
|
|||||||
tbl.add(df)
|
tbl.add(df)
|
||||||
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
||||||
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
||||||
|
|
||||||
|
|
||||||
|
@pytest.mark.slow
|
||||||
|
@pytest.mark.skipif(
|
||||||
|
os.environ.get("WATSONX_API_KEY") is None
|
||||||
|
or os.environ.get("WATSONX_PROJECT_ID") is None,
|
||||||
|
reason="WATSONX_API_KEY and WATSONX_PROJECT_ID not set",
|
||||||
|
)
|
||||||
|
def test_watsonx_embedding(tmp_path):
|
||||||
|
from lancedb.embeddings import WatsonxEmbeddings
|
||||||
|
|
||||||
|
for name in WatsonxEmbeddings.model_names():
|
||||||
|
model = get_registry().get("watsonx").create(max_retries=0, name=name)
|
||||||
|
|
||||||
|
class TextModel(LanceModel):
|
||||||
|
text: str = model.SourceField()
|
||||||
|
vector: Vector(model.ndims()) = model.VectorField()
|
||||||
|
|
||||||
|
db = lancedb.connect("~/.lancedb")
|
||||||
|
tbl = db.create_table("watsonx_test", schema=TextModel, mode="overwrite")
|
||||||
|
df = pd.DataFrame({"text": ["hello world", "goodbye world"]})
|
||||||
|
|
||||||
|
tbl.add(df)
|
||||||
|
assert len(tbl.to_pandas()["vector"][0]) == model.ndims()
|
||||||
|
assert tbl.search("hello").limit(1).to_pandas()["text"][0] == "hello world"
|
||||||
|
|||||||
@@ -42,6 +42,7 @@ async def test_create_scalar_index(some_table: AsyncTable):
|
|||||||
# Can recreate if replace=True
|
# Can recreate if replace=True
|
||||||
await some_table.create_index("id", replace=True)
|
await some_table.create_index("id", replace=True)
|
||||||
indices = await some_table.list_indices()
|
indices = await some_table.list_indices()
|
||||||
|
assert str(indices) == '[Index(BTree, columns=["id"])]'
|
||||||
assert len(indices) == 1
|
assert len(indices) == 1
|
||||||
assert indices[0].index_type == "BTree"
|
assert indices[0].index_type == "BTree"
|
||||||
assert indices[0].columns == ["id"]
|
assert indices[0].columns == ["id"]
|
||||||
|
|||||||
@@ -1,4 +1,5 @@
|
|||||||
import os
|
import os
|
||||||
|
import random
|
||||||
|
|
||||||
import lancedb
|
import lancedb
|
||||||
import numpy as np
|
import numpy as np
|
||||||
@@ -25,10 +26,13 @@ def get_test_table(tmp_path):
|
|||||||
db = lancedb.connect(tmp_path)
|
db = lancedb.connect(tmp_path)
|
||||||
# Create a LanceDB table schema with a vector and a text column
|
# Create a LanceDB table schema with a vector and a text column
|
||||||
emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
||||||
|
meta_emb = EmbeddingFunctionRegistry.get_instance().get("test")()
|
||||||
|
|
||||||
class MyTable(LanceModel):
|
class MyTable(LanceModel):
|
||||||
text: str = emb.SourceField()
|
text: str = emb.SourceField()
|
||||||
vector: Vector(emb.ndims()) = emb.VectorField()
|
vector: Vector(emb.ndims()) = emb.VectorField()
|
||||||
|
meta: str = meta_emb.SourceField()
|
||||||
|
meta_vector: Vector(meta_emb.ndims()) = meta_emb.VectorField()
|
||||||
|
|
||||||
# Initialize the table using the schema
|
# Initialize the table using the schema
|
||||||
table = LanceTable.create(
|
table = LanceTable.create(
|
||||||
@@ -77,7 +81,12 @@ def get_test_table(tmp_path):
|
|||||||
]
|
]
|
||||||
|
|
||||||
# Add the phrases and vectors to the table
|
# Add the phrases and vectors to the table
|
||||||
table.add([{"text": p} for p in phrases])
|
table.add(
|
||||||
|
[
|
||||||
|
{"text": p, "meta": phrases[random.randint(0, len(phrases) - 1)]}
|
||||||
|
for p in phrases
|
||||||
|
]
|
||||||
|
)
|
||||||
|
|
||||||
# Create a fts index
|
# Create a fts index
|
||||||
table.create_fts_index("text")
|
table.create_fts_index("text")
|
||||||
@@ -88,12 +97,12 @@ def get_test_table(tmp_path):
|
|||||||
def _run_test_reranker(reranker, table, query, query_vector, schema):
|
def _run_test_reranker(reranker, table, query, query_vector, schema):
|
||||||
# Hybrid search setting
|
# Hybrid search setting
|
||||||
result1 = (
|
result1 = (
|
||||||
table.search(query, query_type="hybrid")
|
table.search(query, query_type="hybrid", vector_column_name="vector")
|
||||||
.rerank(normalize="score", reranker=reranker)
|
.rerank(normalize="score", reranker=reranker)
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result2 = (
|
result2 = (
|
||||||
table.search(query, query_type="hybrid")
|
table.search(query, query_type="hybrid", vector_column_name="vector")
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
@@ -101,7 +110,7 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
|
|
||||||
query_vector = table.to_pandas()["vector"][0]
|
query_vector = table.to_pandas()["vector"][0]
|
||||||
result = (
|
result = (
|
||||||
table.search((query_vector, query))
|
table.search((query_vector, query), vector_column_name="vector")
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -116,11 +125,16 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
|
|
||||||
# Vector search setting
|
# Vector search setting
|
||||||
result = table.search(query).rerank(reranker=reranker).limit(30).to_arrow()
|
result = (
|
||||||
|
table.search(query, vector_column_name="vector")
|
||||||
|
.rerank(reranker=reranker)
|
||||||
|
.limit(30)
|
||||||
|
.to_arrow()
|
||||||
|
)
|
||||||
assert len(result) == 30
|
assert len(result) == 30
|
||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
result_explicit = (
|
result_explicit = (
|
||||||
table.search(query_vector)
|
table.search(query_vector, vector_column_name="vector")
|
||||||
.rerank(reranker=reranker, query_string=query)
|
.rerank(reranker=reranker, query_string=query)
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -129,11 +143,13 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
with pytest.raises(
|
with pytest.raises(
|
||||||
ValueError
|
ValueError
|
||||||
): # This raises an error because vector query is provided without reanking query
|
): # This raises an error because vector query is provided without reanking query
|
||||||
table.search(query_vector).rerank(reranker=reranker).limit(30).to_arrow()
|
table.search(query_vector, vector_column_name="vector").rerank(
|
||||||
|
reranker=reranker
|
||||||
|
).limit(30).to_arrow()
|
||||||
|
|
||||||
# FTS search setting
|
# FTS search setting
|
||||||
result = (
|
result = (
|
||||||
table.search(query, query_type="fts")
|
table.search(query, query_type="fts", vector_column_name="vector")
|
||||||
.rerank(reranker=reranker)
|
.rerank(reranker=reranker)
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
@@ -141,22 +157,48 @@ def _run_test_reranker(reranker, table, query, query_vector, schema):
|
|||||||
assert len(result) > 0
|
assert len(result) > 0
|
||||||
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
assert np.all(np.diff(result.column("_relevance_score").to_numpy()) <= 0), err
|
||||||
|
|
||||||
|
# Multi-vector search setting
|
||||||
|
rs1 = table.search(query, vector_column_name="vector").limit(10).with_row_id(True)
|
||||||
|
rs2 = (
|
||||||
|
table.search(query, vector_column_name="meta_vector")
|
||||||
|
.limit(10)
|
||||||
|
.with_row_id(True)
|
||||||
|
)
|
||||||
|
result = reranker.rerank_multivector([rs1, rs2], query)
|
||||||
|
assert len(result) == 20
|
||||||
|
result_deduped = reranker.rerank_multivector(
|
||||||
|
[rs1, rs2, rs1], query, deduplicate=True
|
||||||
|
)
|
||||||
|
assert len(result_deduped) < 20
|
||||||
|
result_arrow = reranker.rerank_multivector([rs1.to_arrow(), rs2.to_arrow()], query)
|
||||||
|
assert len(result) == 20 and result == result_arrow
|
||||||
|
|
||||||
|
|
||||||
def _run_test_hybrid_reranker(reranker, tmp_path):
|
def _run_test_hybrid_reranker(reranker, tmp_path):
|
||||||
table, schema = get_test_table(tmp_path)
|
table, schema = get_test_table(tmp_path)
|
||||||
# The default reranker
|
# The default reranker
|
||||||
result1 = (
|
result1 = (
|
||||||
table.search("Our father who art in heaven", query_type="hybrid")
|
table.search(
|
||||||
|
"Our father who art in heaven",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
|
)
|
||||||
.rerank(normalize="score")
|
.rerank(normalize="score")
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result2 = ( # noqa
|
result2 = ( # noqa
|
||||||
table.search("Our father who art in heaven.", query_type="hybrid")
|
table.search(
|
||||||
|
"Our father who art in heaven.",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
|
)
|
||||||
.rerank(normalize="rank")
|
.rerank(normalize="rank")
|
||||||
.to_pydantic(schema)
|
.to_pydantic(schema)
|
||||||
)
|
)
|
||||||
result3 = table.search(
|
result3 = table.search(
|
||||||
"Our father who art in heaven..", query_type="hybrid"
|
"Our father who art in heaven..",
|
||||||
|
query_type="hybrid",
|
||||||
|
vector_column_name="vector",
|
||||||
).to_pydantic(schema)
|
).to_pydantic(schema)
|
||||||
|
|
||||||
assert result1 == result3 # 2 & 3 should be the same as they use score as score
|
assert result1 == result3 # 2 & 3 should be the same as they use score as score
|
||||||
@@ -164,7 +206,7 @@ def _run_test_hybrid_reranker(reranker, tmp_path):
|
|||||||
query = "Our father who art in heaven"
|
query = "Our father who art in heaven"
|
||||||
query_vector = table.to_pandas()["vector"][0]
|
query_vector = table.to_pandas()["vector"][0]
|
||||||
result = (
|
result = (
|
||||||
table.search((query_vector, query))
|
table.search((query_vector, query), vector_column_name="vector")
|
||||||
.limit(30)
|
.limit(30)
|
||||||
.rerank(normalize="score")
|
.rerank(normalize="score")
|
||||||
.to_arrow()
|
.to_arrow()
|
||||||
|
|||||||
@@ -730,7 +730,7 @@ def test_create_scalar_index(db):
|
|||||||
indices = table.to_lance().list_indices()
|
indices = table.to_lance().list_indices()
|
||||||
assert len(indices) == 1
|
assert len(indices) == 1
|
||||||
scalar_index = indices[0]
|
scalar_index = indices[0]
|
||||||
assert scalar_index["type"] == "Scalar"
|
assert scalar_index["type"] == "BTree"
|
||||||
|
|
||||||
# Confirm that prefiltering still works with the scalar index column
|
# Confirm that prefiltering still works with the scalar index column
|
||||||
results = table.search().where("x = 'c'").to_arrow()
|
results = table.search().where("x = 'c'").to_arrow()
|
||||||
@@ -1034,6 +1034,12 @@ async def test_optimize(db_async: AsyncConnection):
|
|||||||
],
|
],
|
||||||
)
|
)
|
||||||
stats = await table.optimize()
|
stats = await table.optimize()
|
||||||
|
expected = (
|
||||||
|
"OptimizeStats(compaction=CompactionStats { fragments_removed: 2, "
|
||||||
|
"fragments_added: 1, files_removed: 2, files_added: 1 }, "
|
||||||
|
"prune=RemovalStats { bytes_removed: 0, old_versions_removed: 0 })"
|
||||||
|
)
|
||||||
|
assert str(stats) == expected
|
||||||
assert stats.compaction.files_removed == 2
|
assert stats.compaction.files_removed == 2
|
||||||
assert stats.compaction.files_added == 1
|
assert stats.compaction.files_added == 1
|
||||||
assert stats.compaction.fragments_added == 1
|
assert stats.compaction.fragments_added == 1
|
||||||
|
|||||||
@@ -1,21 +1,10 @@
|
|||||||
// Copyright 2024 Lance Developers.
|
// SPDX-License-Identifier: Apache-2.0
|
||||||
//
|
// SPDX-FileCopyrightText: Copyright The LanceDB Authors
|
||||||
// Licensed under the Apache License, Version 2.0 (the "License");
|
|
||||||
// you may not use this file except in compliance with the License.
|
|
||||||
// You may obtain a copy of the License at
|
|
||||||
//
|
|
||||||
// http://www.apache.org/licenses/LICENSE-2.0
|
|
||||||
//
|
|
||||||
// Unless required by applicable law or agreed to in writing, software
|
|
||||||
// distributed under the License is distributed on an "AS IS" BASIS,
|
|
||||||
// WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
|
||||||
// See the License for the specific language governing permissions and
|
|
||||||
// limitations under the License.
|
|
||||||
|
|
||||||
use std::{collections::HashMap, sync::Arc, time::Duration};
|
use std::{collections::HashMap, str::FromStr, sync::Arc, time::Duration};
|
||||||
|
|
||||||
use arrow::{datatypes::Schema, ffi_stream::ArrowArrayStreamReader, pyarrow::FromPyArrow};
|
use arrow::{datatypes::Schema, ffi_stream::ArrowArrayStreamReader, pyarrow::FromPyArrow};
|
||||||
use lancedb::connection::{Connection as LanceConnection, CreateTableMode};
|
use lancedb::connection::{Connection as LanceConnection, CreateTableMode, LanceFileVersion};
|
||||||
use pyo3::{
|
use pyo3::{
|
||||||
exceptions::{PyRuntimeError, PyValueError},
|
exceptions::{PyRuntimeError, PyValueError},
|
||||||
pyclass, pyfunction, pymethods, Bound, PyAny, PyRef, PyResult, Python,
|
pyclass, pyfunction, pymethods, Bound, PyAny, PyRef, PyResult, Python,
|
||||||
@@ -91,7 +80,7 @@ impl Connection {
|
|||||||
mode: &str,
|
mode: &str,
|
||||||
data: Bound<'_, PyAny>,
|
data: Bound<'_, PyAny>,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_version: Option<String>,
|
||||||
) -> PyResult<Bound<'a, PyAny>> {
|
) -> PyResult<Bound<'a, PyAny>> {
|
||||||
let inner = self_.get_inner()?.clone();
|
let inner = self_.get_inner()?.clone();
|
||||||
|
|
||||||
@@ -104,8 +93,11 @@ impl Connection {
|
|||||||
builder = builder.storage_options(storage_options);
|
builder = builder.storage_options(storage_options);
|
||||||
}
|
}
|
||||||
|
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_version) = data_storage_version.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_version)
|
||||||
|
.map_err(|e| PyValueError::new_err(e.to_string()))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
|
|
||||||
future_into_py(self_.py(), async move {
|
future_into_py(self_.py(), async move {
|
||||||
@@ -120,7 +112,7 @@ impl Connection {
|
|||||||
mode: &str,
|
mode: &str,
|
||||||
schema: Bound<'_, PyAny>,
|
schema: Bound<'_, PyAny>,
|
||||||
storage_options: Option<HashMap<String, String>>,
|
storage_options: Option<HashMap<String, String>>,
|
||||||
use_legacy_format: Option<bool>,
|
data_storage_version: Option<String>,
|
||||||
) -> PyResult<Bound<'a, PyAny>> {
|
) -> PyResult<Bound<'a, PyAny>> {
|
||||||
let inner = self_.get_inner()?.clone();
|
let inner = self_.get_inner()?.clone();
|
||||||
|
|
||||||
@@ -134,8 +126,11 @@ impl Connection {
|
|||||||
builder = builder.storage_options(storage_options);
|
builder = builder.storage_options(storage_options);
|
||||||
}
|
}
|
||||||
|
|
||||||
if let Some(use_legacy_format) = use_legacy_format {
|
if let Some(data_storage_version) = data_storage_version.as_ref() {
|
||||||
builder = builder.use_legacy_format(use_legacy_format);
|
builder = builder.data_storage_version(
|
||||||
|
LanceFileVersion::from_str(data_storage_version)
|
||||||
|
.map_err(|e| PyValueError::new_err(e.to_string()))?,
|
||||||
|
);
|
||||||
}
|
}
|
||||||
|
|
||||||
future_into_py(self_.py(), async move {
|
future_into_py(self_.py(), async move {
|
||||||
|
|||||||
@@ -98,6 +98,13 @@ pub struct IndexConfig {
|
|||||||
pub columns: Vec<String>,
|
pub columns: Vec<String>,
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[pymethods]
|
||||||
|
impl IndexConfig {
|
||||||
|
pub fn __repr__(&self) -> String {
|
||||||
|
format!("Index({}, columns={:?})", self.index_type, self.columns)
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
impl From<lancedb::index::IndexConfig> for IndexConfig {
|
impl From<lancedb::index::IndexConfig> for IndexConfig {
|
||||||
fn from(value: lancedb::index::IndexConfig) -> Self {
|
fn from(value: lancedb::index::IndexConfig) -> Self {
|
||||||
let index_type = format!("{:?}", value.index_type);
|
let index_type = format!("{:?}", value.index_type);
|
||||||
|
|||||||
@@ -60,6 +60,16 @@ pub struct Table {
|
|||||||
inner: Option<LanceDbTable>,
|
inner: Option<LanceDbTable>,
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[pymethods]
|
||||||
|
impl OptimizeStats {
|
||||||
|
pub fn __repr__(&self) -> String {
|
||||||
|
format!(
|
||||||
|
"OptimizeStats(compaction={:?}, prune={:?})",
|
||||||
|
self.compaction, self.prune
|
||||||
|
)
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
impl Table {
|
impl Table {
|
||||||
pub(crate) fn new(inner: LanceDbTable) -> Self {
|
pub(crate) fn new(inner: LanceDbTable) -> Self {
|
||||||
Self {
|
Self {
|
||||||
@@ -266,6 +276,7 @@ impl Table {
|
|||||||
.optimize(OptimizeAction::Prune {
|
.optimize(OptimizeAction::Prune {
|
||||||
older_than,
|
older_than,
|
||||||
delete_unverified: None,
|
delete_unverified: None,
|
||||||
|
error_if_tagged_old_versions: None,
|
||||||
})
|
})
|
||||||
.await
|
.await
|
||||||
.infer_error()?
|
.infer_error()?
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb-node"
|
name = "lancedb-node"
|
||||||
version = "0.7.2"
|
version = "0.8.0"
|
||||||
description = "Serverless, low-latency vector database for AI applications"
|
description = "Serverless, low-latency vector database for AI applications"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
|
|||||||
@@ -320,12 +320,19 @@ impl JsTable {
|
|||||||
.map(|val| val.value(&mut cx))
|
.map(|val| val.value(&mut cx))
|
||||||
.unwrap_or_default(),
|
.unwrap_or_default(),
|
||||||
);
|
);
|
||||||
|
let error_if_tagged_old_versions: Option<bool> = Some(
|
||||||
|
cx.argument_opt(2)
|
||||||
|
.and_then(|val| val.downcast::<JsBoolean, _>(&mut cx).ok())
|
||||||
|
.map(|val| val.value(&mut cx))
|
||||||
|
.unwrap_or_default(),
|
||||||
|
);
|
||||||
|
|
||||||
rt.spawn(async move {
|
rt.spawn(async move {
|
||||||
let stats = table
|
let stats = table
|
||||||
.optimize(OptimizeAction::Prune {
|
.optimize(OptimizeAction::Prune {
|
||||||
older_than: Some(older_than),
|
older_than: Some(older_than),
|
||||||
delete_unverified,
|
delete_unverified,
|
||||||
|
error_if_tagged_old_versions,
|
||||||
})
|
})
|
||||||
.await;
|
.await;
|
||||||
|
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb"
|
name = "lancedb"
|
||||||
version = "0.7.2"
|
version = "0.8.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
description = "LanceDB: A serverless, low-latency vector database for AI applications"
|
description = "LanceDB: A serverless, low-latency vector database for AI applications"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
@@ -29,6 +29,7 @@ lance-datafusion.workspace = true
|
|||||||
lance-index = { workspace = true }
|
lance-index = { workspace = true }
|
||||||
lance-linalg = { workspace = true }
|
lance-linalg = { workspace = true }
|
||||||
lance-testing = { workspace = true }
|
lance-testing = { workspace = true }
|
||||||
|
lance-encoding = { workspace = true }
|
||||||
pin-project = { workspace = true }
|
pin-project = { workspace = true }
|
||||||
tokio = { version = "1.23", features = ["rt-multi-thread"] }
|
tokio = { version = "1.23", features = ["rt-multi-thread"] }
|
||||||
log.workspace = true
|
log.workspace = true
|
||||||
@@ -46,11 +47,11 @@ serde_with = { version = "3.8.1" }
|
|||||||
reqwest = { version = "0.11.24", features = ["gzip", "json"], optional = true }
|
reqwest = { version = "0.11.24", features = ["gzip", "json"], optional = true }
|
||||||
polars-arrow = { version = ">=0.37,<0.40.0", optional = true }
|
polars-arrow = { version = ">=0.37,<0.40.0", optional = true }
|
||||||
polars = { version = ">=0.37,<0.40.0", optional = true }
|
polars = { version = ">=0.37,<0.40.0", optional = true }
|
||||||
hf-hub = {version = "0.3.2", optional = true}
|
hf-hub = { version = "0.3.2", optional = true }
|
||||||
candle-core = { version = "0.6.0", optional = true }
|
candle-core = { version = "0.6.0", optional = true }
|
||||||
candle-transformers = { version = "0.6.0", optional = true }
|
candle-transformers = { version = "0.6.0", optional = true }
|
||||||
candle-nn = { version = "0.6.0", optional = true }
|
candle-nn = { version = "0.6.0", optional = true }
|
||||||
tokenizers = { version = "0.19.1", optional = true}
|
tokenizers = { version = "0.19.1", optional = true }
|
||||||
|
|
||||||
[dev-dependencies]
|
[dev-dependencies]
|
||||||
tempfile = "3.5.0"
|
tempfile = "3.5.0"
|
||||||
@@ -70,7 +71,13 @@ fp16kernels = ["lance-linalg/fp16kernels"]
|
|||||||
s3-test = []
|
s3-test = []
|
||||||
openai = ["dep:async-openai", "dep:reqwest"]
|
openai = ["dep:async-openai", "dep:reqwest"]
|
||||||
polars = ["dep:polars-arrow", "dep:polars"]
|
polars = ["dep:polars-arrow", "dep:polars"]
|
||||||
sentence-transformers = ["dep:hf-hub", "dep:candle-core", "dep:candle-transformers", "dep:candle-nn", "dep:tokenizers"]
|
sentence-transformers = [
|
||||||
|
"dep:hf-hub",
|
||||||
|
"dep:candle-core",
|
||||||
|
"dep:candle-transformers",
|
||||||
|
"dep:candle-nn",
|
||||||
|
"dep:tokenizers"
|
||||||
|
]
|
||||||
|
|
||||||
[[example]]
|
[[example]]
|
||||||
name = "openai"
|
name = "openai"
|
||||||
|
|||||||
@@ -22,7 +22,7 @@ use std::sync::Arc;
|
|||||||
use arrow_array::{RecordBatchIterator, RecordBatchReader};
|
use arrow_array::{RecordBatchIterator, RecordBatchReader};
|
||||||
use arrow_schema::SchemaRef;
|
use arrow_schema::SchemaRef;
|
||||||
use lance::dataset::{ReadParams, WriteMode};
|
use lance::dataset::{ReadParams, WriteMode};
|
||||||
use lance::io::{ObjectStore, ObjectStoreParams, WrappingObjectStore};
|
use lance::io::{ObjectStore, ObjectStoreParams, ObjectStoreRegistry, WrappingObjectStore};
|
||||||
use object_store::{aws::AwsCredential, local::LocalFileSystem};
|
use object_store::{aws::AwsCredential, local::LocalFileSystem};
|
||||||
use snafu::prelude::*;
|
use snafu::prelude::*;
|
||||||
|
|
||||||
@@ -35,6 +35,7 @@ use crate::io::object_store::MirroringObjectStoreWrapper;
|
|||||||
use crate::table::{NativeTable, TableDefinition, WriteOptions};
|
use crate::table::{NativeTable, TableDefinition, WriteOptions};
|
||||||
use crate::utils::validate_table_name;
|
use crate::utils::validate_table_name;
|
||||||
use crate::Table;
|
use crate::Table;
|
||||||
|
pub use lance_encoding::version::LanceFileVersion;
|
||||||
|
|
||||||
#[cfg(feature = "remote")]
|
#[cfg(feature = "remote")]
|
||||||
use log::warn;
|
use log::warn;
|
||||||
@@ -140,7 +141,7 @@ pub struct CreateTableBuilder<const HAS_DATA: bool, T: IntoArrow> {
|
|||||||
pub(crate) write_options: WriteOptions,
|
pub(crate) write_options: WriteOptions,
|
||||||
pub(crate) table_definition: Option<TableDefinition>,
|
pub(crate) table_definition: Option<TableDefinition>,
|
||||||
pub(crate) embeddings: Vec<(EmbeddingDefinition, Arc<dyn EmbeddingFunction>)>,
|
pub(crate) embeddings: Vec<(EmbeddingDefinition, Arc<dyn EmbeddingFunction>)>,
|
||||||
pub(crate) use_legacy_format: bool,
|
pub(crate) data_storage_version: Option<LanceFileVersion>,
|
||||||
}
|
}
|
||||||
|
|
||||||
// Builder methods that only apply when we have initial data
|
// Builder methods that only apply when we have initial data
|
||||||
@@ -154,7 +155,7 @@ impl<T: IntoArrow> CreateTableBuilder<true, T> {
|
|||||||
write_options: WriteOptions::default(),
|
write_options: WriteOptions::default(),
|
||||||
table_definition: None,
|
table_definition: None,
|
||||||
embeddings: Vec::new(),
|
embeddings: Vec::new(),
|
||||||
use_legacy_format: true,
|
data_storage_version: None,
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
@@ -186,7 +187,7 @@ impl<T: IntoArrow> CreateTableBuilder<true, T> {
|
|||||||
mode: self.mode,
|
mode: self.mode,
|
||||||
write_options: self.write_options,
|
write_options: self.write_options,
|
||||||
embeddings: self.embeddings,
|
embeddings: self.embeddings,
|
||||||
use_legacy_format: self.use_legacy_format,
|
data_storage_version: self.data_storage_version,
|
||||||
};
|
};
|
||||||
Ok((data, builder))
|
Ok((data, builder))
|
||||||
}
|
}
|
||||||
@@ -220,7 +221,7 @@ impl CreateTableBuilder<false, NoData> {
|
|||||||
mode: CreateTableMode::default(),
|
mode: CreateTableMode::default(),
|
||||||
write_options: WriteOptions::default(),
|
write_options: WriteOptions::default(),
|
||||||
embeddings: Vec::new(),
|
embeddings: Vec::new(),
|
||||||
use_legacy_format: true,
|
data_storage_version: None,
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
@@ -283,6 +284,14 @@ impl<const HAS_DATA: bool, T: IntoArrow> CreateTableBuilder<HAS_DATA, T> {
|
|||||||
self
|
self
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/// Set the data storage version.
|
||||||
|
///
|
||||||
|
/// The default is `LanceFileVersion::Legacy`.
|
||||||
|
pub fn data_storage_version(mut self, data_storage_version: LanceFileVersion) -> Self {
|
||||||
|
self.data_storage_version = Some(data_storage_version);
|
||||||
|
self
|
||||||
|
}
|
||||||
|
|
||||||
/// Set to true to use the v1 format for data files
|
/// Set to true to use the v1 format for data files
|
||||||
///
|
///
|
||||||
/// This is currently defaulted to true and can be set to false to opt-in
|
/// This is currently defaulted to true and can be set to false to opt-in
|
||||||
@@ -292,8 +301,13 @@ impl<const HAS_DATA: bool, T: IntoArrow> CreateTableBuilder<HAS_DATA, T> {
|
|||||||
///
|
///
|
||||||
/// Once the new format is stable, the default will change to `false` for
|
/// Once the new format is stable, the default will change to `false` for
|
||||||
/// several releases and then eventually this option will be removed.
|
/// several releases and then eventually this option will be removed.
|
||||||
|
#[deprecated(since = "0.9.0", note = "use data_storage_version instead")]
|
||||||
pub fn use_legacy_format(mut self, use_legacy_format: bool) -> Self {
|
pub fn use_legacy_format(mut self, use_legacy_format: bool) -> Self {
|
||||||
self.use_legacy_format = use_legacy_format;
|
self.data_storage_version = if use_legacy_format {
|
||||||
|
Some(LanceFileVersion::Legacy)
|
||||||
|
} else {
|
||||||
|
Some(LanceFileVersion::Stable)
|
||||||
|
};
|
||||||
self
|
self
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
@@ -789,13 +803,14 @@ impl Database {
|
|||||||
|
|
||||||
let plain_uri = url.to_string();
|
let plain_uri = url.to_string();
|
||||||
|
|
||||||
|
let registry = Arc::new(ObjectStoreRegistry::default());
|
||||||
let storage_options = options.storage_options.clone();
|
let storage_options = options.storage_options.clone();
|
||||||
let os_params = ObjectStoreParams {
|
let os_params = ObjectStoreParams {
|
||||||
storage_options: Some(storage_options.clone()),
|
storage_options: Some(storage_options.clone()),
|
||||||
..Default::default()
|
..Default::default()
|
||||||
};
|
};
|
||||||
let (object_store, base_path) =
|
let (object_store, base_path) =
|
||||||
ObjectStore::from_uri_and_params(&plain_uri, &os_params).await?;
|
ObjectStore::from_uri_and_params(registry, &plain_uri, &os_params).await?;
|
||||||
if object_store.is_local() {
|
if object_store.is_local() {
|
||||||
Self::try_create_dir(&plain_uri).context(CreateDirSnafu { path: plain_uri })?;
|
Self::try_create_dir(&plain_uri).context(CreateDirSnafu { path: plain_uri })?;
|
||||||
}
|
}
|
||||||
@@ -961,7 +976,7 @@ impl ConnectionInternal for Database {
|
|||||||
if matches!(&options.mode, CreateTableMode::Overwrite) {
|
if matches!(&options.mode, CreateTableMode::Overwrite) {
|
||||||
write_params.mode = WriteMode::Overwrite;
|
write_params.mode = WriteMode::Overwrite;
|
||||||
}
|
}
|
||||||
write_params.use_legacy_format = options.use_legacy_format;
|
write_params.data_storage_version = options.data_storage_version;
|
||||||
|
|
||||||
match NativeTable::create(
|
match NativeTable::create(
|
||||||
&table_uri,
|
&table_uri,
|
||||||
|
|||||||
@@ -191,6 +191,8 @@ pub enum OptimizeAction {
|
|||||||
/// Because they may be part of an in-progress transaction, files newer than 7 days old are not deleted by default.
|
/// Because they may be part of an in-progress transaction, files newer than 7 days old are not deleted by default.
|
||||||
/// If you are sure that there are no in-progress transactions, then you can set this to True to delete all files older than `older_than`.
|
/// If you are sure that there are no in-progress transactions, then you can set this to True to delete all files older than `older_than`.
|
||||||
delete_unverified: Option<bool>,
|
delete_unverified: Option<bool>,
|
||||||
|
/// If true, an error will be returned if there are any old versions that are still tagged.
|
||||||
|
error_if_tagged_old_versions: Option<bool>,
|
||||||
},
|
},
|
||||||
/// Optimize the indices
|
/// Optimize the indices
|
||||||
///
|
///
|
||||||
@@ -1079,8 +1081,8 @@ impl NativeTable {
|
|||||||
params: Option<WriteParams>,
|
params: Option<WriteParams>,
|
||||||
read_consistency_interval: Option<std::time::Duration>,
|
read_consistency_interval: Option<std::time::Duration>,
|
||||||
) -> Result<Self> {
|
) -> Result<Self> {
|
||||||
|
// Default params uses format v1.
|
||||||
let params = params.unwrap_or(WriteParams {
|
let params = params.unwrap_or(WriteParams {
|
||||||
use_legacy_format: true,
|
|
||||||
..Default::default()
|
..Default::default()
|
||||||
});
|
});
|
||||||
// patch the params if we have a write store wrapper
|
// patch the params if we have a write store wrapper
|
||||||
@@ -1173,12 +1175,13 @@ impl NativeTable {
|
|||||||
&self,
|
&self,
|
||||||
older_than: Duration,
|
older_than: Duration,
|
||||||
delete_unverified: Option<bool>,
|
delete_unverified: Option<bool>,
|
||||||
|
error_if_tagged_old_versions: Option<bool>,
|
||||||
) -> Result<RemovalStats> {
|
) -> Result<RemovalStats> {
|
||||||
Ok(self
|
Ok(self
|
||||||
.dataset
|
.dataset
|
||||||
.get_mut()
|
.get_mut()
|
||||||
.await?
|
.await?
|
||||||
.cleanup_old_versions(older_than, delete_unverified)
|
.cleanup_old_versions(older_than, delete_unverified, error_if_tagged_old_versions)
|
||||||
.await?)
|
.await?)
|
||||||
}
|
}
|
||||||
|
|
||||||
@@ -1506,8 +1509,8 @@ impl NativeTable {
|
|||||||
}
|
}
|
||||||
|
|
||||||
let mut dataset = self.dataset.get_mut().await?;
|
let mut dataset = self.dataset.get_mut().await?;
|
||||||
let lance_idx_params = lance::index::scalar::ScalarIndexParams {
|
let lance_idx_params = lance_index::scalar::ScalarIndexParams {
|
||||||
force_index_type: Some(lance::index::scalar::ScalarIndexType::BTree),
|
force_index_type: Some(lance_index::scalar::ScalarIndexType::BTree),
|
||||||
};
|
};
|
||||||
dataset
|
dataset
|
||||||
.create_index(
|
.create_index(
|
||||||
@@ -1607,6 +1610,9 @@ impl TableInternal for NativeTable {
|
|||||||
let data =
|
let data =
|
||||||
MaybeEmbedded::try_new(data, self.table_definition().await?, add.embedding_registry)?;
|
MaybeEmbedded::try_new(data, self.table_definition().await?, add.embedding_registry)?;
|
||||||
|
|
||||||
|
// Still use the legacy lance format (v1) by default.
|
||||||
|
// We don't want to accidentally switch to v2 format during an add operation.
|
||||||
|
// If the table is already v2 this won't have any effect.
|
||||||
let mut lance_params = add.write_options.lance_write_params.unwrap_or(WriteParams {
|
let mut lance_params = add.write_options.lance_write_params.unwrap_or(WriteParams {
|
||||||
mode: match add.mode {
|
mode: match add.mode {
|
||||||
AddDataMode::Append => WriteMode::Append,
|
AddDataMode::Append => WriteMode::Append,
|
||||||
@@ -1628,16 +1634,11 @@ impl TableInternal for NativeTable {
|
|||||||
}
|
}
|
||||||
|
|
||||||
// patch the params if we have a write store wrapper
|
// patch the params if we have a write store wrapper
|
||||||
let mut lance_params = match self.store_wrapper.clone() {
|
let lance_params = match self.store_wrapper.clone() {
|
||||||
Some(wrapper) => lance_params.patch_with_store_wrapper(wrapper)?,
|
Some(wrapper) => lance_params.patch_with_store_wrapper(wrapper)?,
|
||||||
None => lance_params,
|
None => lance_params,
|
||||||
};
|
};
|
||||||
|
|
||||||
// Only use the new format if the user passes use_legacy_format=False in while creating
|
|
||||||
// a table with data. We don't want to accidentally switch to v2 format during an add
|
|
||||||
// operation. If the table is already v2 this won't have any effect.
|
|
||||||
lance_params.use_legacy_format = true;
|
|
||||||
|
|
||||||
self.dataset.ensure_mutable().await?;
|
self.dataset.ensure_mutable().await?;
|
||||||
let dataset = Dataset::write(data, &self.uri, Some(lance_params)).await?;
|
let dataset = Dataset::write(data, &self.uri, Some(lance_params)).await?;
|
||||||
|
|
||||||
@@ -1878,6 +1879,7 @@ impl TableInternal for NativeTable {
|
|||||||
.optimize(OptimizeAction::Prune {
|
.optimize(OptimizeAction::Prune {
|
||||||
older_than: None,
|
older_than: None,
|
||||||
delete_unverified: None,
|
delete_unverified: None,
|
||||||
|
error_if_tagged_old_versions: None,
|
||||||
})
|
})
|
||||||
.await?
|
.await?
|
||||||
.prune;
|
.prune;
|
||||||
@@ -1893,11 +1895,13 @@ impl TableInternal for NativeTable {
|
|||||||
OptimizeAction::Prune {
|
OptimizeAction::Prune {
|
||||||
older_than,
|
older_than,
|
||||||
delete_unverified,
|
delete_unverified,
|
||||||
|
error_if_tagged_old_versions,
|
||||||
} => {
|
} => {
|
||||||
stats.prune = Some(
|
stats.prune = Some(
|
||||||
self.cleanup_old_versions(
|
self.cleanup_old_versions(
|
||||||
older_than.unwrap_or(Duration::try_days(7).expect("valid delta")),
|
older_than.unwrap_or(Duration::try_days(7).expect("valid delta")),
|
||||||
delete_unverified,
|
delete_unverified,
|
||||||
|
error_if_tagged_old_versions,
|
||||||
)
|
)
|
||||||
.await?,
|
.await?,
|
||||||
);
|
);
|
||||||
|
|||||||
Reference in New Issue
Block a user