mirror of
https://github.com/lancedb/lancedb.git
synced 2025-12-23 13:29:57 +00:00
Compare commits
21 Commits
python-v0.
...
python-v0.
| Author | SHA1 | Date | |
|---|---|---|---|
|
|
38f0031d0b | ||
|
|
e118c37228 | ||
|
|
abeaae3d80 | ||
|
|
b3c0227065 | ||
|
|
521e665f57 | ||
|
|
ffb28dd4fc | ||
|
|
32af962c0c | ||
|
|
18484d0b6c | ||
|
|
c02ee3c80c | ||
|
|
dcd5f51036 | ||
|
|
9b8472850e | ||
|
|
36d05ea641 | ||
|
|
7ed86cadfb | ||
|
|
1c123b58d8 | ||
|
|
bf7d2d6fb0 | ||
|
|
c7732585bf | ||
|
|
b3bf6386c3 | ||
|
|
4b79db72bf | ||
|
|
622a2922e2 | ||
|
|
c91221d710 | ||
|
|
56da5ebd13 |
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.10.0-beta.1"
|
current_version = "0.10.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
@@ -24,34 +24,56 @@ commit = true
|
|||||||
message = "Bump version: {current_version} → {new_version}"
|
message = "Bump version: {current_version} → {new_version}"
|
||||||
commit_args = ""
|
commit_args = ""
|
||||||
|
|
||||||
|
# Java maven files
|
||||||
|
pre_commit_hooks = [
|
||||||
|
"""
|
||||||
|
NEW_VERSION="${BVHOOK_NEW_MAJOR}.${BVHOOK_NEW_MINOR}.${BVHOOK_NEW_PATCH}"
|
||||||
|
if [ ! -z "$BVHOOK_NEW_PRE_L" ] && [ ! -z "$BVHOOK_NEW_PRE_N" ]; then
|
||||||
|
NEW_VERSION="${NEW_VERSION}-${BVHOOK_NEW_PRE_L}.${BVHOOK_NEW_PRE_N}"
|
||||||
|
fi
|
||||||
|
echo "Constructed new version: $NEW_VERSION"
|
||||||
|
cd java && mvn versions:set -DnewVersion=$NEW_VERSION && mvn versions:commit
|
||||||
|
|
||||||
|
# Check for any modified but unstaged pom.xml files
|
||||||
|
MODIFIED_POMS=$(git ls-files -m | grep pom.xml)
|
||||||
|
if [ ! -z "$MODIFIED_POMS" ]; then
|
||||||
|
echo "The following pom.xml files were modified but not staged. Adding them now:"
|
||||||
|
echo "$MODIFIED_POMS" | while read -r file; do
|
||||||
|
git add "$file"
|
||||||
|
echo "Added: $file"
|
||||||
|
done
|
||||||
|
fi
|
||||||
|
""",
|
||||||
|
]
|
||||||
|
|
||||||
[tool.bumpversion.parts.pre_l]
|
[tool.bumpversion.parts.pre_l]
|
||||||
values = ["beta", "final"]
|
|
||||||
optional_value = "final"
|
optional_value = "final"
|
||||||
|
values = ["beta", "final"]
|
||||||
|
|
||||||
[[tool.bumpversion.files]]
|
[[tool.bumpversion.files]]
|
||||||
filename = "node/package.json"
|
filename = "node/package.json"
|
||||||
search = "\"version\": \"{current_version}\","
|
|
||||||
replace = "\"version\": \"{new_version}\","
|
replace = "\"version\": \"{new_version}\","
|
||||||
|
search = "\"version\": \"{current_version}\","
|
||||||
|
|
||||||
[[tool.bumpversion.files]]
|
[[tool.bumpversion.files]]
|
||||||
filename = "nodejs/package.json"
|
filename = "nodejs/package.json"
|
||||||
search = "\"version\": \"{current_version}\","
|
|
||||||
replace = "\"version\": \"{new_version}\","
|
replace = "\"version\": \"{new_version}\","
|
||||||
|
search = "\"version\": \"{current_version}\","
|
||||||
|
|
||||||
# nodejs binary packages
|
# nodejs binary packages
|
||||||
[[tool.bumpversion.files]]
|
[[tool.bumpversion.files]]
|
||||||
glob = "nodejs/npm/*/package.json"
|
glob = "nodejs/npm/*/package.json"
|
||||||
search = "\"version\": \"{current_version}\","
|
|
||||||
replace = "\"version\": \"{new_version}\","
|
replace = "\"version\": \"{new_version}\","
|
||||||
|
search = "\"version\": \"{current_version}\","
|
||||||
|
|
||||||
# Cargo files
|
# Cargo files
|
||||||
# ------------
|
# ------------
|
||||||
[[tool.bumpversion.files]]
|
[[tool.bumpversion.files]]
|
||||||
filename = "rust/ffi/node/Cargo.toml"
|
filename = "rust/ffi/node/Cargo.toml"
|
||||||
search = "\nversion = \"{current_version}\""
|
|
||||||
replace = "\nversion = \"{new_version}\""
|
replace = "\nversion = \"{new_version}\""
|
||||||
|
search = "\nversion = \"{current_version}\""
|
||||||
|
|
||||||
[[tool.bumpversion.files]]
|
[[tool.bumpversion.files]]
|
||||||
filename = "rust/lancedb/Cargo.toml"
|
filename = "rust/lancedb/Cargo.toml"
|
||||||
search = "\nversion = \"{current_version}\""
|
|
||||||
replace = "\nversion = \"{new_version}\""
|
replace = "\nversion = \"{new_version}\""
|
||||||
|
search = "\nversion = \"{current_version}\""
|
||||||
|
|||||||
5
.github/workflows/java-publish.yml
vendored
5
.github/workflows/java-publish.yml
vendored
@@ -94,11 +94,16 @@ jobs:
|
|||||||
mkdir -p ./core/target/classes/nativelib/darwin-aarch64 ./core/target/classes/nativelib/linux-aarch64
|
mkdir -p ./core/target/classes/nativelib/darwin-aarch64 ./core/target/classes/nativelib/linux-aarch64
|
||||||
cp ../liblancedb_jni_darwin_aarch64.zip/liblancedb_jni.dylib ./core/target/classes/nativelib/darwin-aarch64/liblancedb_jni.dylib
|
cp ../liblancedb_jni_darwin_aarch64.zip/liblancedb_jni.dylib ./core/target/classes/nativelib/darwin-aarch64/liblancedb_jni.dylib
|
||||||
cp ../liblancedb_jni_linux_aarch64.zip/liblancedb_jni.so ./core/target/classes/nativelib/linux-aarch64/liblancedb_jni.so
|
cp ../liblancedb_jni_linux_aarch64.zip/liblancedb_jni.so ./core/target/classes/nativelib/linux-aarch64/liblancedb_jni.so
|
||||||
|
- name: Dry run
|
||||||
|
if: github.event_name == 'pull_request'
|
||||||
|
run: |
|
||||||
|
mvn --batch-mode -DskipTests package
|
||||||
- name: Set github
|
- name: Set github
|
||||||
run: |
|
run: |
|
||||||
git config --global user.email "LanceDB Github Runner"
|
git config --global user.email "LanceDB Github Runner"
|
||||||
git config --global user.name "dev+gha@lancedb.com"
|
git config --global user.name "dev+gha@lancedb.com"
|
||||||
- name: Publish with Java 8
|
- name: Publish with Java 8
|
||||||
|
if: github.event_name == 'release'
|
||||||
run: |
|
run: |
|
||||||
echo "use-agent" >> ~/.gnupg/gpg.conf
|
echo "use-agent" >> ~/.gnupg/gpg.conf
|
||||||
echo "pinentry-mode loopback" >> ~/.gnupg/gpg.conf
|
echo "pinentry-mode loopback" >> ~/.gnupg/gpg.conf
|
||||||
|
|||||||
2
.github/workflows/make-release-commit.yml
vendored
2
.github/workflows/make-release-commit.yml
vendored
@@ -30,7 +30,7 @@ on:
|
|||||||
default: true
|
default: true
|
||||||
type: boolean
|
type: boolean
|
||||||
other:
|
other:
|
||||||
description: 'Make a Node/Rust release'
|
description: 'Make a Node/Rust/Java release'
|
||||||
required: true
|
required: true
|
||||||
default: true
|
default: true
|
||||||
type: boolean
|
type: boolean
|
||||||
|
|||||||
14
Cargo.toml
14
Cargo.toml
@@ -20,13 +20,13 @@ keywords = ["lancedb", "lance", "database", "vector", "search"]
|
|||||||
categories = ["database-implementations"]
|
categories = ["database-implementations"]
|
||||||
|
|
||||||
[workspace.dependencies]
|
[workspace.dependencies]
|
||||||
lance = { "version" = "=0.17.0", "features" = ["dynamodb"] }
|
lance = { "version" = "=0.18.0", "features" = ["dynamodb"] }
|
||||||
lance-index = { "version" = "=0.17.0" }
|
lance-index = { "version" = "=0.18.0" }
|
||||||
lance-linalg = { "version" = "=0.17.0" }
|
lance-linalg = { "version" = "=0.18.0" }
|
||||||
lance-table = { "version" = "=0.17.0" }
|
lance-table = { "version" = "=0.18.0" }
|
||||||
lance-testing = { "version" = "=0.17.0" }
|
lance-testing = { "version" = "=0.18.0" }
|
||||||
lance-datafusion = { "version" = "=0.17.0" }
|
lance-datafusion = { "version" = "=0.18.0" }
|
||||||
lance-encoding = { "version" = "=0.17.0" }
|
lance-encoding = { "version" = "=0.18.0" }
|
||||||
# Note that this one does not include pyarrow
|
# Note that this one does not include pyarrow
|
||||||
arrow = { version = "52.2", optional = false }
|
arrow = { version = "52.2", optional = false }
|
||||||
arrow-array = "52.2"
|
arrow-array = "52.2"
|
||||||
|
|||||||
@@ -106,6 +106,17 @@ nav:
|
|||||||
- Overview: hybrid_search/hybrid_search.md
|
- Overview: hybrid_search/hybrid_search.md
|
||||||
- Comparing Rerankers: hybrid_search/eval.md
|
- Comparing Rerankers: hybrid_search/eval.md
|
||||||
- Airbnb financial data example: notebooks/hybrid_search.ipynb
|
- Airbnb financial data example: notebooks/hybrid_search.ipynb
|
||||||
|
- RAG:
|
||||||
|
- Vanilla RAG: rag/vanilla_rag.md
|
||||||
|
- Multi-head RAG: rag/multi_head_rag.md
|
||||||
|
- Corrective RAG: rag/corrective_rag.md
|
||||||
|
- Agentic RAG: rag/agentic_rag.md
|
||||||
|
- Graph RAG: rag/graph_rag.md
|
||||||
|
- Self RAG: rag/self_rag.md
|
||||||
|
- Adaptive RAG: rag/adaptive_rag.md
|
||||||
|
- Advanced Techniques:
|
||||||
|
- HyDE: rag/advanced_techniques/hyde.md
|
||||||
|
- FLARE: rag/advanced_techniques/flare.md
|
||||||
- Reranking:
|
- Reranking:
|
||||||
- Quickstart: reranking/index.md
|
- Quickstart: reranking/index.md
|
||||||
- Cohere Reranker: reranking/cohere.md
|
- Cohere Reranker: reranking/cohere.md
|
||||||
@@ -127,7 +138,8 @@ nav:
|
|||||||
- Reranking: guides/tuning_retrievers/2_reranking.md
|
- Reranking: guides/tuning_retrievers/2_reranking.md
|
||||||
- Embedding fine-tuning: guides/tuning_retrievers/3_embed_tuning.md
|
- Embedding fine-tuning: guides/tuning_retrievers/3_embed_tuning.md
|
||||||
- 🧬 Managing embeddings:
|
- 🧬 Managing embeddings:
|
||||||
- Overview: embeddings/index.md
|
- Understand Embeddings: embeddings/understanding_embeddings.md
|
||||||
|
- Get Started: embeddings/index.md
|
||||||
- Embedding functions: embeddings/embedding_functions.md
|
- Embedding functions: embeddings/embedding_functions.md
|
||||||
- Available models:
|
- Available models:
|
||||||
- Overview: embeddings/default_embedding_functions.md
|
- Overview: embeddings/default_embedding_functions.md
|
||||||
@@ -220,6 +232,17 @@ nav:
|
|||||||
- Overview: hybrid_search/hybrid_search.md
|
- Overview: hybrid_search/hybrid_search.md
|
||||||
- Comparing Rerankers: hybrid_search/eval.md
|
- Comparing Rerankers: hybrid_search/eval.md
|
||||||
- Airbnb financial data example: notebooks/hybrid_search.ipynb
|
- Airbnb financial data example: notebooks/hybrid_search.ipynb
|
||||||
|
- RAG:
|
||||||
|
- Vanilla RAG: rag/vanilla_rag.md
|
||||||
|
- Multi-head RAG: rag/multi_head_rag.md
|
||||||
|
- Corrective RAG: rag/corrective_rag.md
|
||||||
|
- Agentic RAG: rag/agentic_rag.md
|
||||||
|
- Graph RAG: rag/graph_rag.md
|
||||||
|
- Self RAG: rag/self_rag.md
|
||||||
|
- Adaptive RAG: rag/adaptive_rag.md
|
||||||
|
- Advanced Techniques:
|
||||||
|
- HyDE: rag/advanced_techniques/hyde.md
|
||||||
|
- FLARE: rag/advanced_techniques/flare.md
|
||||||
- Reranking:
|
- Reranking:
|
||||||
- Quickstart: reranking/index.md
|
- Quickstart: reranking/index.md
|
||||||
- Cohere Reranker: reranking/cohere.md
|
- Cohere Reranker: reranking/cohere.md
|
||||||
@@ -241,7 +264,8 @@ nav:
|
|||||||
- Reranking: guides/tuning_retrievers/2_reranking.md
|
- Reranking: guides/tuning_retrievers/2_reranking.md
|
||||||
- Embedding fine-tuning: guides/tuning_retrievers/3_embed_tuning.md
|
- Embedding fine-tuning: guides/tuning_retrievers/3_embed_tuning.md
|
||||||
- Managing Embeddings:
|
- Managing Embeddings:
|
||||||
- Overview: embeddings/index.md
|
- Understand Embeddings: embeddings/understanding_embeddings.md
|
||||||
|
- Get Started: embeddings/index.md
|
||||||
- Embedding functions: embeddings/embedding_functions.md
|
- Embedding functions: embeddings/embedding_functions.md
|
||||||
- Available models:
|
- Available models:
|
||||||
- Overview: embeddings/default_embedding_functions.md
|
- Overview: embeddings/default_embedding_functions.md
|
||||||
|
|||||||
133
docs/src/embeddings/understanding_embeddings.md
Normal file
133
docs/src/embeddings/understanding_embeddings.md
Normal file
@@ -0,0 +1,133 @@
|
|||||||
|
# Understand Embeddings
|
||||||
|
|
||||||
|
The term **dimension** is a synonym for the number of elements in a feature vector. Each feature can be thought of as a different axis in a geometric space.
|
||||||
|
|
||||||
|
High-dimensional data means there are many features(or attributes) in the data.
|
||||||
|
|
||||||
|
!!! example
|
||||||
|
1. An image is a data point and it might have thousands of dimensions because each pixel could be considered as a feature.
|
||||||
|
|
||||||
|
2. Text data, when represented by each word or character, can also lead to high dimensions, especially when considering all possible words in a language.
|
||||||
|
|
||||||
|
Embedding captures **meaning and relationships** within data by mapping high-dimensional data into a lower-dimensional space. It captures it by placing inputs that are more **similar in meaning** closer together in the **embedding space**.
|
||||||
|
|
||||||
|
## What are Vector Embeddings?
|
||||||
|
|
||||||
|
Vector embeddings is a way to convert complex data, like text, images, or audio into numerical coordinates (called vectors) that can be plotted in an n-dimensional space(embedding space).
|
||||||
|
|
||||||
|
The closer these data points are related in the real world, the closer their corresponding numerical coordinates (vectors) will be to each other in the embedding space. This proximity in the embedding space reflects their semantic similarities, allowing machines to intuitively understand and process the data in a way that mirrors human perception of relationships and meaning.
|
||||||
|
|
||||||
|
In a way, it captures the most important aspects of the data while ignoring the less important ones. As a result, tasks like searching for related content or identifying patterns become more efficient and accurate, as the embeddings make it possible to quantify how **closely related** different **data points** are and **reduce** the **computational complexity**.
|
||||||
|
|
||||||
|
??? question "Are vectors and embeddings the same thing?"
|
||||||
|
|
||||||
|
When we say “vectors” we mean - **list of numbers** that **represents the data**.
|
||||||
|
When we say “embeddings” we mean - **list of numbers** that **capture important details and relationships**.
|
||||||
|
|
||||||
|
Although the terms are often used interchangeably, “embeddings” highlight how the data is represented with meaning and structure, while “vector” simply refers to the numerical form of that representation.
|
||||||
|
|
||||||
|
## Embedding vs Indexing
|
||||||
|
|
||||||
|
We already saw that creating **embeddings** on data is a method of creating **vectors** for a **n-dimensional embedding space** that captures the meaning and relationships inherent in the data.
|
||||||
|
|
||||||
|
Once we have these **vectors**, indexing comes into play. Indexing is a method of organizing these vector embeddings, that allows us to quickly and efficiently locate and retrieve them from the entire dataset of vector embeddings.
|
||||||
|
|
||||||
|
## What types of data/objects can be embedded?
|
||||||
|
|
||||||
|
The following are common types of data that can be embedded:
|
||||||
|
|
||||||
|
1. **Text**: Text data includes sentences, paragraphs, documents, or any written content.
|
||||||
|
2. **Images**: Image data encompasses photographs, illustrations, or any visual content.
|
||||||
|
3. **Audio**: Audio data includes sounds, music, speech, or any auditory content.
|
||||||
|
4. **Video**: Video data consists of moving images and sound, which can convey complex information.
|
||||||
|
|
||||||
|
Large datasets of multi-modal data (text, audio, images, etc.) can be converted into embeddings with the appropriate model.
|
||||||
|
|
||||||
|
!!! tip "LanceDB vs Other traditional Vector DBs"
|
||||||
|
While many vector databases primarily focus on the storage and retrieval of vector embeddings, **LanceDB** uses **Lance file format** (operates on a disk-based architecture), which allows for the storage and management of not just embeddings but also **raw file data (bytes)**. This capability means that users can integrate various types of data, including images and text, alongside their vector embeddings in a unified system.
|
||||||
|
|
||||||
|
With the ability to store both vectors and associated file data, LanceDB enhances the querying process. Users can perform semantic searches that not only retrieve similar embeddings but also access related files and metadata, thus streamlining the workflow.
|
||||||
|
|
||||||
|
## How does embedding works?
|
||||||
|
|
||||||
|
As mentioned, after creating embedding, each data point is represented as a vector in a n-dimensional space (embedding space). The dimensionality of this space can vary depending on the complexity of the data and the specific embedding technique used.
|
||||||
|
|
||||||
|
Points that are close to each other in vector space are considered similar (or appear in similar contexts), and points that are far away are considered dissimilar. To quantify this closeness, we use distance as a metric which can be measured in the following way -
|
||||||
|
|
||||||
|
1. **Euclidean Distance (L2)**: It calculates the straight-line distance between two points (vectors) in a multidimensional space.
|
||||||
|
2. **Cosine Similarity**: It measures the cosine of the angle between two vectors, providing a normalized measure of similarity based on their direction.
|
||||||
|
3. **Dot product**: It is calculated as the sum of the products of their corresponding components. To measure relatedness it considers both the magnitude and direction of the vectors.
|
||||||
|
|
||||||
|
## How do you create and store vector embeddings for your data?
|
||||||
|
|
||||||
|
1. **Creating embeddings**: Choose an embedding model, it can be a pre-trained model (open-source or commercial) or you can train a custom embedding model for your scenario. Then feed your preprocessed data into the chosen model to obtain embeddings.
|
||||||
|
|
||||||
|
??? question "Popular choices for embedding models"
|
||||||
|
For text data, popular choices are OpenAI’s text-embedding models, Google Gemini text-embedding models, Cohere’s Embed models, and SentenceTransformers, etc.
|
||||||
|
|
||||||
|
For image data, popular choices are CLIP (Contrastive Language–Image Pretraining), Imagebind embeddings by meta (supports audio, video, and image), and Jina multi-modal embeddings, etc.
|
||||||
|
|
||||||
|
2. **Storing vector embeddings**: This effectively requires **specialized databases** that can handle the complexity of vector data, as traditional databases often struggle with this task. Vector databases are designed specifically for storing and querying vector embeddings. They optimize for efficient nearest-neighbor searches and provide built-in indexing mechanisms.
|
||||||
|
|
||||||
|
!!! tip "Why LanceDB"
|
||||||
|
LanceDB **automates** the entire process of creating and storing embeddings for your data. LanceDB allows you to define and use **embedding functions**, which can be **pre-trained models** or **custom models**.
|
||||||
|
|
||||||
|
This enables you to **generate** embeddings tailored to the nature of your data (e.g., text, images) and **store** both the **original data** and **embeddings** in a **structured schema** thus providing efficient querying capabilities for similarity searches.
|
||||||
|
|
||||||
|
Let's quickly [get started](./index.md) and learn how to manage embeddings in LanceDB.
|
||||||
|
|
||||||
|
## Bonus: As a developer, what you can create using embeddings?
|
||||||
|
|
||||||
|
As a developer, you can create a variety of innovative applications using vector embeddings. Check out the following -
|
||||||
|
|
||||||
|
<div class="grid cards" markdown>
|
||||||
|
|
||||||
|
- __Chatbots__
|
||||||
|
|
||||||
|
---
|
||||||
|
|
||||||
|
Develop chatbots that utilize embeddings to retrieve relevant context and generate coherent, contextually aware responses to user queries.
|
||||||
|
|
||||||
|
[:octicons-arrow-right-24: Check out examples](../examples/python_examples/chatbot.md)
|
||||||
|
|
||||||
|
- __Recommendation Systems__
|
||||||
|
|
||||||
|
---
|
||||||
|
|
||||||
|
Develop systems that recommend content (such as articles, movies, or products) based on the similarity of keywords and descriptions, enhancing user experience.
|
||||||
|
|
||||||
|
[:octicons-arrow-right-24: Check out examples](../examples/python_examples/recommendersystem.md)
|
||||||
|
|
||||||
|
- __Vector Search__
|
||||||
|
|
||||||
|
---
|
||||||
|
|
||||||
|
Build powerful applications that harness the full potential of semantic search, enabling them to retrieve relevant data quickly and effectively.
|
||||||
|
|
||||||
|
[:octicons-arrow-right-24: Check out examples](../examples/python_examples/vector_search.md)
|
||||||
|
|
||||||
|
- __RAG Applications__
|
||||||
|
|
||||||
|
---
|
||||||
|
|
||||||
|
Combine the strengths of large language models (LLMs) with retrieval-based approaches to create more useful applications.
|
||||||
|
|
||||||
|
[:octicons-arrow-right-24: Check out examples](../examples/python_examples/rag.md)
|
||||||
|
|
||||||
|
- __Many more examples__
|
||||||
|
|
||||||
|
---
|
||||||
|
|
||||||
|
Explore applied examples available as Colab notebooks or Python scripts to integrate into your applications.
|
||||||
|
|
||||||
|
[:octicons-arrow-right-24: More](../examples/examples_python.md)
|
||||||
|
|
||||||
|
</div>
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
|
|
||||||
@@ -2,7 +2,7 @@
|
|||||||
|
|
||||||
LanceDB provides support for full-text search via Lance (before via [Tantivy](https://github.com/quickwit-oss/tantivy) (Python only)), allowing you to incorporate keyword-based search (based on BM25) in your retrieval solutions.
|
LanceDB provides support for full-text search via Lance (before via [Tantivy](https://github.com/quickwit-oss/tantivy) (Python only)), allowing you to incorporate keyword-based search (based on BM25) in your retrieval solutions.
|
||||||
|
|
||||||
Currently, the Lance full text search is missing some features that are in the Tantivy full text search. This includes phrase queries, re-ranking, and customizing the tokenizer. Thus, in Python, Tantivy is still the default way to do full text search and many of the instructions below apply just to Tantivy-based indices.
|
Currently, the Lance full text search is missing some features that are in the Tantivy full text search. This includes query parser and customizing the tokenizer. Thus, in Python, Tantivy is still the default way to do full text search and many of the instructions below apply just to Tantivy-based indices.
|
||||||
|
|
||||||
|
|
||||||
## Installation (Only for Tantivy-based FTS)
|
## Installation (Only for Tantivy-based FTS)
|
||||||
@@ -205,7 +205,7 @@ table.create_fts_index(["text_field"], use_tantivy=True, ordering_field_names=["
|
|||||||
## Phrase queries vs. terms queries
|
## Phrase queries vs. terms queries
|
||||||
|
|
||||||
!!! warning "Warn"
|
!!! warning "Warn"
|
||||||
Lance-based FTS doesn't support queries combining by boolean operators `OR`, `AND`.
|
Lance-based FTS doesn't support queries using boolean operators `OR`, `AND`.
|
||||||
|
|
||||||
For full-text search you can specify either a **phrase** query like `"the old man and the sea"`,
|
For full-text search you can specify either a **phrase** query like `"the old man and the sea"`,
|
||||||
or a **terms** search query like `"(Old AND Man) AND Sea"`. For more details on the terms
|
or a **terms** search query like `"(Old AND Man) AND Sea"`. For more details on the terms
|
||||||
|
|||||||
@@ -68,3 +68,25 @@ currently is also a memory intensive operation.
|
|||||||
#### Returns
|
#### Returns
|
||||||
|
|
||||||
[`Index`](Index.md)
|
[`Index`](Index.md)
|
||||||
|
|
||||||
|
### fts()
|
||||||
|
|
||||||
|
> `static` **fts**(`options`?): [`Index`](Index.md)
|
||||||
|
|
||||||
|
Create a full text search index
|
||||||
|
|
||||||
|
This index is used to search for text data. The index is created by tokenizing the text
|
||||||
|
into words and then storing occurrences of these words in a data structure called inverted index
|
||||||
|
that allows for fast search.
|
||||||
|
|
||||||
|
During a search the query is tokenized and the inverted index is used to find the rows that
|
||||||
|
contain the query words. The rows are then scored based on BM25 and the top scoring rows are
|
||||||
|
sorted and returned.
|
||||||
|
|
||||||
|
#### Parameters
|
||||||
|
|
||||||
|
• **options?**: `Partial`<[`FtsOptions`](../interfaces/FtsOptions.md)>
|
||||||
|
|
||||||
|
#### Returns
|
||||||
|
|
||||||
|
[`Index`](Index.md)
|
||||||
|
|||||||
@@ -501,16 +501,28 @@ Get the schema of the table.
|
|||||||
|
|
||||||
#### search(query)
|
#### search(query)
|
||||||
|
|
||||||
> `abstract` **search**(`query`): [`VectorQuery`](VectorQuery.md)
|
> `abstract` **search**(`query`, `queryType`, `ftsColumns`): [`VectorQuery`](VectorQuery.md)
|
||||||
|
|
||||||
Create a search query to find the nearest neighbors
|
Create a search query to find the nearest neighbors
|
||||||
of the given query vector
|
of the given query vector, or the documents
|
||||||
|
with the highest relevance to the query string.
|
||||||
|
|
||||||
##### Parameters
|
##### Parameters
|
||||||
|
|
||||||
• **query**: `string`
|
• **query**: `string`
|
||||||
|
|
||||||
the query. This will be converted to a vector using the table's provided embedding function
|
the query. This will be converted to a vector using the table's provided embedding function,
|
||||||
|
or the query string for full-text search if `queryType` is "fts".
|
||||||
|
|
||||||
|
• **queryType**: `string` = `"auto"` \| `"fts"`
|
||||||
|
|
||||||
|
the type of query to run. If "auto", the query type will be determined based on the query.
|
||||||
|
|
||||||
|
• **ftsColumns**: `string[] | str` = undefined
|
||||||
|
|
||||||
|
the columns to search in. If not provided, all indexed columns will be searched.
|
||||||
|
|
||||||
|
For now, this can support to search only one column.
|
||||||
|
|
||||||
##### Returns
|
##### Returns
|
||||||
|
|
||||||
|
|||||||
@@ -37,6 +37,7 @@
|
|||||||
- [IndexOptions](interfaces/IndexOptions.md)
|
- [IndexOptions](interfaces/IndexOptions.md)
|
||||||
- [IndexStatistics](interfaces/IndexStatistics.md)
|
- [IndexStatistics](interfaces/IndexStatistics.md)
|
||||||
- [IvfPqOptions](interfaces/IvfPqOptions.md)
|
- [IvfPqOptions](interfaces/IvfPqOptions.md)
|
||||||
|
- [FtsOptions](interfaces/FtsOptions.md)
|
||||||
- [TableNamesOptions](interfaces/TableNamesOptions.md)
|
- [TableNamesOptions](interfaces/TableNamesOptions.md)
|
||||||
- [UpdateOptions](interfaces/UpdateOptions.md)
|
- [UpdateOptions](interfaces/UpdateOptions.md)
|
||||||
- [WriteOptions](interfaces/WriteOptions.md)
|
- [WriteOptions](interfaces/WriteOptions.md)
|
||||||
|
|||||||
51
docs/src/rag/adaptive_rag.md
Normal file
51
docs/src/rag/adaptive_rag.md
Normal file
@@ -0,0 +1,51 @@
|
|||||||
|
**Adaptive RAG 🤹♂️**
|
||||||
|
====================================================================
|
||||||
|
Adaptive RAG introduces a RAG technique that combines query analysis with self-corrective RAG.
|
||||||
|
|
||||||
|
For Query Analysis, it uses a small classifier(LLM), to decide the query’s complexity. Query Analysis helps routing smoothly to adjust between different retrieval strategies No retrieval, Single-shot RAG or Iterative RAG.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/pdf/2403.14403)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>Adaptive-RAG: <a href="https://github.com/starsuzi/Adaptive-RAG">Source</a>
|
||||||
|
</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
**[Offical Implementation](https://github.com/starsuzi/Adaptive-RAG)**
|
||||||
|
|
||||||
|
Here’s a code snippet for query analysis
|
||||||
|
|
||||||
|
```python
|
||||||
|
from langchain_core.prompts import ChatPromptTemplate
|
||||||
|
from langchain_core.pydantic_v1 import BaseModel, Field
|
||||||
|
from langchain_openai import ChatOpenAI
|
||||||
|
|
||||||
|
class RouteQuery(BaseModel):
|
||||||
|
"""Route a user query to the most relevant datasource."""
|
||||||
|
|
||||||
|
datasource: Literal["vectorstore", "web_search"] = Field(
|
||||||
|
...,
|
||||||
|
description="Given a user question choose to route it to web search or a vectorstore.",
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
# LLM with function call
|
||||||
|
llm = ChatOpenAI(model="gpt-3.5-turbo-0125", temperature=0)
|
||||||
|
structured_llm_router = llm.with_structured_output(RouteQuery)
|
||||||
|
```
|
||||||
|
|
||||||
|
For defining and querying retriever
|
||||||
|
|
||||||
|
```python
|
||||||
|
# add documents in LanceDB
|
||||||
|
vectorstore = LanceDB.from_documents(
|
||||||
|
documents=doc_splits,
|
||||||
|
embedding=OpenAIEmbeddings(),
|
||||||
|
)
|
||||||
|
retriever = vectorstore.as_retriever()
|
||||||
|
|
||||||
|
# query using defined retriever
|
||||||
|
question = "How adaptive RAG works"
|
||||||
|
docs = retriever.get_relevant_documents(question)
|
||||||
|
```
|
||||||
38
docs/src/rag/advanced_techniques/flare.md
Normal file
38
docs/src/rag/advanced_techniques/flare.md
Normal file
@@ -0,0 +1,38 @@
|
|||||||
|
**FLARE 💥**
|
||||||
|
====================================================================
|
||||||
|
FLARE, stands for Forward-Looking Active REtrieval augmented generation is a generic retrieval-augmented generation method that actively decides when and what to retrieve using a prediction of the upcoming sentence to anticipate future content and utilize it as the query to retrieve relevant documents if it contains low-confidence tokens.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/abs/2305.06983)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>FLARE: <a href="https://github.com/jzbjyb/FLARE">Source</a></figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR/main.ipynb)
|
||||||
|
|
||||||
|
Here’s a code snippet for using FLARE with Langchain
|
||||||
|
|
||||||
|
```python
|
||||||
|
from langchain.vectorstores import LanceDB
|
||||||
|
from langchain.document_loaders import ArxivLoader
|
||||||
|
from langchain.chains import FlareChain
|
||||||
|
from langchain.prompts import PromptTemplate
|
||||||
|
from langchain.chains import LLMChain
|
||||||
|
from langchain.llms import OpenAI
|
||||||
|
|
||||||
|
llm = OpenAI()
|
||||||
|
|
||||||
|
# load dataset
|
||||||
|
|
||||||
|
# LanceDB retriever
|
||||||
|
vector_store = LanceDB.from_documents(doc_chunks, embeddings, connection=table)
|
||||||
|
retriever = vector_store.as_retriever()
|
||||||
|
|
||||||
|
# define flare chain
|
||||||
|
flare = FlareChain.from_llm(llm=llm,retriever=vector_store_retriever,max_generation_len=300,min_prob=0.45)
|
||||||
|
|
||||||
|
result = flare.run(input_text)
|
||||||
|
```
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/better-rag-FLAIR/main.ipynb)
|
||||||
55
docs/src/rag/advanced_techniques/hyde.md
Normal file
55
docs/src/rag/advanced_techniques/hyde.md
Normal file
@@ -0,0 +1,55 @@
|
|||||||
|
**HyDE: Hypothetical Document Embeddings 🤹♂️**
|
||||||
|
====================================================================
|
||||||
|
HyDE, stands for Hypothetical Document Embeddings is an approach used for precise zero-shot dense retrieval without relevance labels. It focuses on augmenting and improving similarity searches, often intertwined with vector stores in information retrieval. The method generates a hypothetical document for an incoming query, which is then embedded and used to look up real documents that are similar to the hypothetical document.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/pdf/2212.10496)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>HyDE: <a href="https://arxiv.org/pdf/2212.10496">Source</a></figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE/main.ipynb)
|
||||||
|
|
||||||
|
Here’s a code snippet for using HyDE with Langchain
|
||||||
|
|
||||||
|
```python
|
||||||
|
from langchain.llms import OpenAI
|
||||||
|
from langchain.embeddings import OpenAIEmbeddings
|
||||||
|
from langchain.prompts import PromptTemplate
|
||||||
|
from langchain.chains import LLMChain, HypotheticalDocumentEmbedder
|
||||||
|
from langchain.vectorstores import LanceDB
|
||||||
|
|
||||||
|
# set OPENAI_API_KEY as env variable before this step
|
||||||
|
# initialize LLM and embedding function
|
||||||
|
llm = OpenAI()
|
||||||
|
emebeddings = OpenAIEmbeddings()
|
||||||
|
|
||||||
|
# HyDE embedding
|
||||||
|
embeddings = HypotheticalDocumentEmbedder(llm_chain=llm_chain,base_embeddings=embeddings)
|
||||||
|
|
||||||
|
# load dataset
|
||||||
|
|
||||||
|
# LanceDB retriever
|
||||||
|
retriever = LanceDB.from_documents(documents, embeddings, connection=table)
|
||||||
|
|
||||||
|
# prompt template
|
||||||
|
prompt_template = """
|
||||||
|
As a knowledgeable and helpful research assistant, your task is to provide informative answers based on the given context. Use your extensive knowledge base to offer clear, concise, and accurate responses to the user's inquiries.
|
||||||
|
if quetion is not related to documents simply say you dont know
|
||||||
|
Question: {question}
|
||||||
|
|
||||||
|
Answer:
|
||||||
|
"""
|
||||||
|
|
||||||
|
prompt = PromptTemplate(input_variables=["question"], template=prompt_template)
|
||||||
|
|
||||||
|
# LLM Chain
|
||||||
|
llm_chain = LLMChain(llm=llm, prompt=prompt)
|
||||||
|
|
||||||
|
# vector search
|
||||||
|
retriever.similarity_search(query)
|
||||||
|
llm_chain.run(query)
|
||||||
|
```
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Advance-RAG-with-HyDE/main.ipynb)
|
||||||
101
docs/src/rag/agentic_rag.md
Normal file
101
docs/src/rag/agentic_rag.md
Normal file
@@ -0,0 +1,101 @@
|
|||||||
|
**Agentic RAG 🤖**
|
||||||
|
====================================================================
|
||||||
|
Agentic RAG is Agent-based RAG introduces an advanced framework for answering questions by using intelligent agents instead of just relying on large language models. These agents act like expert researchers, handling complex tasks such as detailed planning, multi-step reasoning, and using external tools. They navigate multiple documents, compare information, and generate accurate answers. This system is easily scalable, with each new document set managed by a sub-agent, making it a powerful tool for tackling a wide range of information needs.
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|
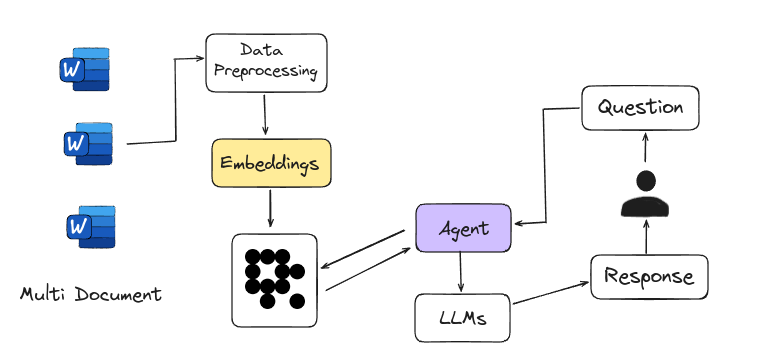
|
||||||
|
<figcaption>Agent-based RAG</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG/main.ipynb)
|
||||||
|
|
||||||
|
Here’s a code snippet for defining retriever using Langchain
|
||||||
|
|
||||||
|
```python
|
||||||
|
from langchain.text_splitter import RecursiveCharacterTextSplitter
|
||||||
|
from langchain_community.document_loaders import WebBaseLoader
|
||||||
|
from langchain_community.vectorstores import LanceDB
|
||||||
|
from langchain_openai import OpenAIEmbeddings
|
||||||
|
|
||||||
|
urls = [
|
||||||
|
"https://content.dgft.gov.in/Website/CIEP.pdf",
|
||||||
|
"https://content.dgft.gov.in/Website/GAE.pdf",
|
||||||
|
"https://content.dgft.gov.in/Website/HTE.pdf",
|
||||||
|
]
|
||||||
|
|
||||||
|
|
||||||
|
docs = [WebBaseLoader(url).load() for url in urls]
|
||||||
|
docs_list = [item for sublist in docs for item in sublist]
|
||||||
|
|
||||||
|
text_splitter = RecursiveCharacterTextSplitter.from_tiktoken_encoder(
|
||||||
|
chunk_size=100, chunk_overlap=50
|
||||||
|
)
|
||||||
|
doc_splits = text_splitter.split_documents(docs_list)
|
||||||
|
|
||||||
|
# add documents in LanceDB
|
||||||
|
vectorstore = LanceDB.from_documents(
|
||||||
|
documents=doc_splits,
|
||||||
|
embedding=OpenAIEmbeddings(),
|
||||||
|
)
|
||||||
|
retriever = vectorstore.as_retriever()
|
||||||
|
|
||||||
|
```
|
||||||
|
|
||||||
|
Agent that formulates an improved query for better retrieval results and then grades the retrieved documents
|
||||||
|
|
||||||
|
```python
|
||||||
|
def grade_documents(state) -> Literal["generate", "rewrite"]:
|
||||||
|
class grade(BaseModel):
|
||||||
|
binary_score: str = Field(description="Relevance score 'yes' or 'no'")
|
||||||
|
|
||||||
|
model = ChatOpenAI(temperature=0, model="gpt-4-0125-preview", streaming=True)
|
||||||
|
llm_with_tool = model.with_structured_output(grade)
|
||||||
|
prompt = PromptTemplate(
|
||||||
|
template="""You are a grader assessing relevance of a retrieved document to a user question. \n
|
||||||
|
Here is the retrieved document: \n\n {context} \n\n
|
||||||
|
Here is the user question: {question} \n
|
||||||
|
If the document contains keyword(s) or semantic meaning related to the user question, grade it as relevant. \n
|
||||||
|
Give a binary score 'yes' or 'no' score to indicate whether the document is relevant to the question.""",
|
||||||
|
input_variables=["context", "question"],
|
||||||
|
)
|
||||||
|
chain = prompt | llm_with_tool
|
||||||
|
|
||||||
|
messages = state["messages"]
|
||||||
|
last_message = messages[-1]
|
||||||
|
question = messages[0].content
|
||||||
|
docs = last_message.content
|
||||||
|
|
||||||
|
scored_result = chain.invoke({"question": question, "context": docs})
|
||||||
|
score = scored_result.binary_score
|
||||||
|
|
||||||
|
return "generate" if score == "yes" else "rewrite"
|
||||||
|
|
||||||
|
|
||||||
|
def agent(state):
|
||||||
|
messages = state["messages"]
|
||||||
|
model = ChatOpenAI(temperature=0, streaming=True, model="gpt-4-turbo")
|
||||||
|
model = model.bind_tools(tools)
|
||||||
|
response = model.invoke(messages)
|
||||||
|
return {"messages": [response]}
|
||||||
|
|
||||||
|
|
||||||
|
def rewrite(state):
|
||||||
|
messages = state["messages"]
|
||||||
|
question = messages[0].content
|
||||||
|
msg = [
|
||||||
|
HumanMessage(
|
||||||
|
content=f""" \n
|
||||||
|
Look at the input and try to reason about the underlying semantic intent / meaning. \n
|
||||||
|
Here is the initial question:
|
||||||
|
\n ------- \n
|
||||||
|
{question}
|
||||||
|
\n ------- \n
|
||||||
|
Formulate an improved question: """,
|
||||||
|
)
|
||||||
|
]
|
||||||
|
model = ChatOpenAI(temperature=0, model="gpt-4-0125-preview", streaming=True)
|
||||||
|
response = model.invoke(msg)
|
||||||
|
return {"messages": [response]}
|
||||||
|
```
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Agentic_RAG/main.ipynb)
|
||||||
120
docs/src/rag/corrective_rag.md
Normal file
120
docs/src/rag/corrective_rag.md
Normal file
@@ -0,0 +1,120 @@
|
|||||||
|
**Corrective RAG ✅**
|
||||||
|
====================================================================
|
||||||
|
|
||||||
|
Corrective-RAG (CRAG) is a strategy for Retrieval-Augmented Generation (RAG) that includes self-reflection and self-grading of retrieved documents. Here’s a simplified breakdown of the steps involved:
|
||||||
|
|
||||||
|
1. **Relevance Check**: If at least one document meets the relevance threshold, the process moves forward to the generation phase.
|
||||||
|
2. **Knowledge Refinement**: Before generating an answer, the process refines the knowledge by dividing the document into smaller segments called "knowledge strips."
|
||||||
|
3. **Grading and Filtering**: Each "knowledge strip" is graded, and irrelevant ones are filtered out.
|
||||||
|
4. **Additional Data Source**: If all documents are below the relevance threshold, or if the system is unsure about their relevance, it will seek additional information by performing a web search to supplement the retrieved data.
|
||||||
|
|
||||||
|
Above steps are mentioned in
|
||||||
|
**[Official Paper](https://arxiv.org/abs/2401.15884)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>Corrective RAG: <a href="https://github.com/HuskyInSalt/CRAG">Source</a>
|
||||||
|
</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
Corrective Retrieval-Augmented Generation (CRAG) is a method that works like a **built-in fact-checker**.
|
||||||
|
|
||||||
|
**[Offical Implementation](https://github.com/HuskyInSalt/CRAG)**
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph/CRAG_with_Langgraph.ipynb)
|
||||||
|
|
||||||
|
Here’s a code snippet for defining a table with the [Embedding API](https://lancedb.github.io/lancedb/embeddings/embedding_functions/), and retrieves the relevant documents.
|
||||||
|
|
||||||
|
```python
|
||||||
|
import pandas as pd
|
||||||
|
import lancedb
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.embeddings import get_registry
|
||||||
|
|
||||||
|
db = lancedb.connect("/tmp/db")
|
||||||
|
model = get_registry().get("sentence-transformers").create(name="BAAI/bge-small-en-v1.5", device="cpu")
|
||||||
|
|
||||||
|
class Docs(LanceModel):
|
||||||
|
text: str = model.SourceField()
|
||||||
|
vector: Vector(model.ndims()) = model.VectorField()
|
||||||
|
|
||||||
|
table = db.create_table("docs", schema=Docs)
|
||||||
|
|
||||||
|
# considering chunks are in list format
|
||||||
|
df = pd.DataFrame({'text':chunks})
|
||||||
|
table.add(data=df)
|
||||||
|
|
||||||
|
# as per document feeded
|
||||||
|
query = "How Transformers work?"
|
||||||
|
actual = table.search(query).limit(1).to_list()[0]
|
||||||
|
print(actual.text)
|
||||||
|
```
|
||||||
|
|
||||||
|
Code snippet for grading retrieved documents, filtering out irrelevant ones, and performing a web search if necessary:
|
||||||
|
|
||||||
|
```python
|
||||||
|
def grade_documents(state):
|
||||||
|
"""
|
||||||
|
Determines whether the retrieved documents are relevant to the question
|
||||||
|
|
||||||
|
Args:
|
||||||
|
state (dict): The current graph state
|
||||||
|
|
||||||
|
Returns:
|
||||||
|
state (dict): Updates documents key with relevant documents
|
||||||
|
"""
|
||||||
|
|
||||||
|
state_dict = state["keys"]
|
||||||
|
question = state_dict["question"]
|
||||||
|
documents = state_dict["documents"]
|
||||||
|
|
||||||
|
class grade(BaseModel):
|
||||||
|
"""
|
||||||
|
Binary score for relevance check
|
||||||
|
"""
|
||||||
|
|
||||||
|
binary_score: str = Field(description="Relevance score 'yes' or 'no'")
|
||||||
|
|
||||||
|
model = ChatOpenAI(temperature=0, model="gpt-4-0125-preview", streaming=True)
|
||||||
|
# grading using openai
|
||||||
|
grade_tool_oai = convert_to_openai_tool(grade)
|
||||||
|
llm_with_tool = model.bind(
|
||||||
|
tools=[convert_to_openai_tool(grade_tool_oai)],
|
||||||
|
tool_choice={"type": "function", "function": {"name": "grade"}},
|
||||||
|
)
|
||||||
|
|
||||||
|
parser_tool = PydanticToolsParser(tools=[grade])
|
||||||
|
prompt = PromptTemplate(
|
||||||
|
template="""You are a grader assessing relevance of a retrieved document to a user question. \n
|
||||||
|
Here is the retrieved document: \n\n {context} \n\n
|
||||||
|
Here is the user question: {question} \n
|
||||||
|
If the document contains keyword(s) or semantic meaning related to the user question, grade it as relevant. \n
|
||||||
|
Give a binary score 'yes' or 'no' score to indicate whether the document is relevant to the question.""",
|
||||||
|
input_variables=["context", "question"],
|
||||||
|
)
|
||||||
|
|
||||||
|
chain = prompt | llm_with_tool | parser_tool
|
||||||
|
|
||||||
|
filtered_docs = []
|
||||||
|
search = "No"
|
||||||
|
for d in documents:
|
||||||
|
score = chain.invoke({"question": question, "context": d.page_content})
|
||||||
|
grade = score[0].binary_score
|
||||||
|
if grade == "yes":
|
||||||
|
filtered_docs.append(d)
|
||||||
|
else:
|
||||||
|
search = "Yes"
|
||||||
|
continue
|
||||||
|
|
||||||
|
return {
|
||||||
|
"keys": {
|
||||||
|
"documents": filtered_docs,
|
||||||
|
"question": question,
|
||||||
|
"run_web_search": search,
|
||||||
|
}
|
||||||
|
}
|
||||||
|
```
|
||||||
|
|
||||||
|
Check Colab for the Implementation of CRAG with Langgraph
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/Corrective-RAG-with_Langgraph/CRAG_with_Langgraph.ipynb)
|
||||||
54
docs/src/rag/graph_rag.md
Normal file
54
docs/src/rag/graph_rag.md
Normal file
@@ -0,0 +1,54 @@
|
|||||||
|
**Graph RAG 📊**
|
||||||
|
====================================================================
|
||||||
|
Graph RAG uses knowledge graphs together with large language models (LLMs) to improve how information is retrieved and generated. It overcomes the limits of traditional search methods by using knowledge graphs, which organize data as connected entities and relationships.
|
||||||
|
|
||||||
|
One of the main benefits of Graph RAG is its ability to capture and represent complex relationships between entities, something that traditional text-based retrieval systems struggle with. By using this structured knowledge, LLMs can better grasp the context and details of a query, resulting in more accurate and insightful answers.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/pdf/2404.16130)**
|
||||||
|
|
||||||
|
**[Offical Implementation](https://github.com/microsoft/graphrag)**
|
||||||
|
|
||||||
|
[Microsoft Research Blog](https://www.microsoft.com/en-us/research/blog/graphrag-unlocking-llm-discovery-on-narrative-private-data/)
|
||||||
|
|
||||||
|
!!! note "Default VectorDB"
|
||||||
|
|
||||||
|
Graph RAG uses LanceDB as the default vector database for performing vector search to retrieve relevant entities.
|
||||||
|
|
||||||
|
Working with Graph RAG is quite straightforward
|
||||||
|
|
||||||
|
- **Installation and API KEY as env variable**
|
||||||
|
|
||||||
|
Set `OPENAI_API_KEY` as `GRAPHRAG_API_KEY`
|
||||||
|
|
||||||
|
```bash
|
||||||
|
pip install graphrag
|
||||||
|
export GRAPHRAG_API_KEY="sk-..."
|
||||||
|
```
|
||||||
|
|
||||||
|
- **Initial structure for indexing dataset**
|
||||||
|
|
||||||
|
```bash
|
||||||
|
python3 -m graphrag.index --init --root dataset-dir
|
||||||
|
```
|
||||||
|
|
||||||
|
- **Index Dataset**
|
||||||
|
|
||||||
|
```bash
|
||||||
|
python3 -m graphrag.index --root dataset-dir
|
||||||
|
```
|
||||||
|
|
||||||
|
- **Execute Query**
|
||||||
|
|
||||||
|
Global Query Execution gives a broad overview of dataset
|
||||||
|
|
||||||
|
```bash
|
||||||
|
python3 -m graphrag.query --root dataset-dir --method global "query-question"
|
||||||
|
```
|
||||||
|
|
||||||
|
Local Query Execution gives a detailed and specific answers based on the context of the entities
|
||||||
|
|
||||||
|
```bash
|
||||||
|
python3 -m graphrag.query --root dataset-dir --method local "query-question"
|
||||||
|
```
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/examples/Graphrag/main.ipynb)
|
||||||
49
docs/src/rag/multi_head_rag.md
Normal file
49
docs/src/rag/multi_head_rag.md
Normal file
@@ -0,0 +1,49 @@
|
|||||||
|
**Multi-Head RAG 📃**
|
||||||
|
====================================================================
|
||||||
|
|
||||||
|
Multi-head RAG (MRAG) is designed to handle queries that need multiple documents with diverse content. These queries are tough because the documents’ embeddings can be far apart, making retrieval difficult. MRAG simplifies this by using the activations from a Transformer's multi-head attention layer, rather than the decoder layer, to fetch these varied documents. Different attention heads capture different aspects of the data, so using these activations helps create embeddings that better represent various data facets and improves retrieval accuracy for complex queries.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/pdf/2406.05085)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>Multi-Head RAG: <a href="https://github.com/spcl/MRAG">Source</a>
|
||||||
|
</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
MRAG is cost-effective and energy-efficient because it avoids extra LLM queries, multiple model instances, increased storage, and additional inference passes.
|
||||||
|
|
||||||
|
**[Official Implementation](https://github.com/spcl/MRAG)**
|
||||||
|
|
||||||
|
Here’s a code snippet for defining different embedding spaces with the [Embedding API](https://lancedb.github.io/lancedb/embeddings/embedding_functions/)
|
||||||
|
|
||||||
|
```python
|
||||||
|
import lancedb
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.embeddings import get_registry
|
||||||
|
|
||||||
|
# model definition using LanceDB Embedding API
|
||||||
|
model1 = get_registry().get("openai").create()
|
||||||
|
model2 = get_registry().get("ollama").create(name="llama3")
|
||||||
|
model3 = get_registry().get("ollama").create(name="mistral")
|
||||||
|
|
||||||
|
|
||||||
|
# define schema for creating embedding spaces with Embedding API
|
||||||
|
class Space1(LanceModel):
|
||||||
|
text: str = model1.SourceField()
|
||||||
|
vector: Vector(model1.ndims()) = model1.VectorField()
|
||||||
|
|
||||||
|
|
||||||
|
class Space2(LanceModel):
|
||||||
|
text: str = model2.SourceField()
|
||||||
|
vector: Vector(model2.ndims()) = model2.VectorField()
|
||||||
|
|
||||||
|
|
||||||
|
class Space3(LanceModel):
|
||||||
|
text: str = model3.SourceField()
|
||||||
|
vector: Vector(model3.ndims()) = model3.VectorField()
|
||||||
|
```
|
||||||
|
|
||||||
|
Create different tables using defined embedding spaces, then make queries to each embedding space. Use the resulted closest documents from each embedding space to generate answers.
|
||||||
|
|
||||||
|
|
||||||
96
docs/src/rag/self_rag.md
Normal file
96
docs/src/rag/self_rag.md
Normal file
@@ -0,0 +1,96 @@
|
|||||||
|
**Self RAG 🤳**
|
||||||
|
====================================================================
|
||||||
|
Self-RAG is a strategy for Retrieval-Augmented Generation (RAG) to get better retrieved information, generated text, and checking their own work, all without losing their flexibility. Unlike the traditional Retrieval-Augmented Generation (RAG) method, Self-RAG retrieves information as needed, can skip retrieval if not needed, and evaluates its own output while generating text. It also uses a process to pick the best output based on different preferences.
|
||||||
|
|
||||||
|
**[Official Paper](https://arxiv.org/pdf/2310.11511)**
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>Self RAG: <a href="https://github.com/AkariAsai/self-rag">Source</a>
|
||||||
|
</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
**[Offical Implementation](https://github.com/AkariAsai/self-rag)**
|
||||||
|
|
||||||
|
Self-RAG starts by generating a response without retrieving extra info if it's not needed. For questions that need more details, it retrieves to get the necessary information.
|
||||||
|
|
||||||
|
Here’s a code snippet for defining retriever using Langchain
|
||||||
|
|
||||||
|
```python
|
||||||
|
from langchain.text_splitter import RecursiveCharacterTextSplitter
|
||||||
|
from langchain_community.document_loaders import WebBaseLoader
|
||||||
|
from langchain_community.vectorstores import LanceDB
|
||||||
|
from langchain_openai import OpenAIEmbeddings
|
||||||
|
|
||||||
|
urls = [
|
||||||
|
"https://lilianweng.github.io/posts/2023-06-23-agent/",
|
||||||
|
"https://lilianweng.github.io/posts/2023-03-15-prompt-engineering/",
|
||||||
|
"https://lilianweng.github.io/posts/2023-10-25-adv-attack-llm/",
|
||||||
|
]
|
||||||
|
|
||||||
|
|
||||||
|
docs = [WebBaseLoader(url).load() for url in urls]
|
||||||
|
docs_list = [item for sublist in docs for item in sublist]
|
||||||
|
|
||||||
|
text_splitter = RecursiveCharacterTextSplitter.from_tiktoken_encoder(
|
||||||
|
chunk_size=100, chunk_overlap=50
|
||||||
|
)
|
||||||
|
doc_splits = text_splitter.split_documents(docs_list)
|
||||||
|
|
||||||
|
# add documents in LanceDB
|
||||||
|
vectorstore = LanceDB.from_documents(
|
||||||
|
documents=doc_splits,
|
||||||
|
embedding=OpenAIEmbeddings(),
|
||||||
|
)
|
||||||
|
retriever = vectorstore.as_retriever()
|
||||||
|
|
||||||
|
```
|
||||||
|
|
||||||
|
Functions that grades the retrieved documents and if required formulates an improved query for better retrieval results
|
||||||
|
|
||||||
|
```python
|
||||||
|
def grade_documents(state) -> Literal["generate", "rewrite"]:
|
||||||
|
class grade(BaseModel):
|
||||||
|
binary_score: str = Field(description="Relevance score 'yes' or 'no'")
|
||||||
|
|
||||||
|
model = ChatOpenAI(temperature=0, model="gpt-4-0125-preview", streaming=True)
|
||||||
|
llm_with_tool = model.with_structured_output(grade)
|
||||||
|
prompt = PromptTemplate(
|
||||||
|
template="""You are a grader assessing relevance of a retrieved document to a user question. \n
|
||||||
|
Here is the retrieved document: \n\n {context} \n\n
|
||||||
|
Here is the user question: {question} \n
|
||||||
|
If the document contains keyword(s) or semantic meaning related to the user question, grade it as relevant. \n
|
||||||
|
Give a binary score 'yes' or 'no' score to indicate whether the document is relevant to the question.""",
|
||||||
|
input_variables=["context", "question"],
|
||||||
|
)
|
||||||
|
chain = prompt | llm_with_tool
|
||||||
|
|
||||||
|
messages = state["messages"]
|
||||||
|
last_message = messages[-1]
|
||||||
|
question = messages[0].content
|
||||||
|
docs = last_message.content
|
||||||
|
|
||||||
|
scored_result = chain.invoke({"question": question, "context": docs})
|
||||||
|
score = scored_result.binary_score
|
||||||
|
|
||||||
|
return "generate" if score == "yes" else "rewrite"
|
||||||
|
|
||||||
|
|
||||||
|
def rewrite(state):
|
||||||
|
messages = state["messages"]
|
||||||
|
question = messages[0].content
|
||||||
|
msg = [
|
||||||
|
HumanMessage(
|
||||||
|
content=f""" \n
|
||||||
|
Look at the input and try to reason about the underlying semantic intent / meaning. \n
|
||||||
|
Here is the initial question:
|
||||||
|
\n ------- \n
|
||||||
|
{question}
|
||||||
|
\n ------- \n
|
||||||
|
Formulate an improved question: """,
|
||||||
|
)
|
||||||
|
]
|
||||||
|
model = ChatOpenAI(temperature=0, model="gpt-4-0125-preview", streaming=True)
|
||||||
|
response = model.invoke(msg)
|
||||||
|
return {"messages": [response]}
|
||||||
|
```
|
||||||
54
docs/src/rag/vanilla_rag.md
Normal file
54
docs/src/rag/vanilla_rag.md
Normal file
@@ -0,0 +1,54 @@
|
|||||||
|
**Vanilla RAG 🌱**
|
||||||
|
====================================================================
|
||||||
|
|
||||||
|
RAG(Retrieval-Augmented Generation) works by finding documents related to the user's question, combining them with a prompt for a large language model (LLM), and then using the LLM to create more accurate and relevant answers.
|
||||||
|
|
||||||
|
Here’s a simple guide to building a RAG pipeline from scratch:
|
||||||
|
|
||||||
|
1. **Data Loading**: Gather and load the documents you want to use for answering questions.
|
||||||
|
|
||||||
|
2. **Chunking and Embedding**: Split the documents into smaller chunks and convert them into numerical vectors (embeddings) that capture their meaning.
|
||||||
|
|
||||||
|
3. **Vector Store**: Create a LanceDB table to store and manage these vectors for quick access during retrieval.
|
||||||
|
|
||||||
|
4. **Retrieval & Prompt Preparation**: When a question is asked, find the most relevant document chunks from the table and prepare a prompt combining these chunks with the question.
|
||||||
|
|
||||||
|
5. **Answer Generation**: Send the prepared prompt to a LLM to generate a detailed and accurate answer.
|
||||||
|
|
||||||
|
<figure markdown="span">
|
||||||
|

|
||||||
|
<figcaption>Vanilla RAG
|
||||||
|
</figcaption>
|
||||||
|
</figure>
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/RAG-from-Scratch/RAG_from_Scratch.ipynb)
|
||||||
|
|
||||||
|
Here’s a code snippet for defining a table with the [Embedding API](https://lancedb.github.io/lancedb/embeddings/embedding_functions/), which simplifies the process by handling embedding extraction and querying in one step.
|
||||||
|
|
||||||
|
```python
|
||||||
|
import pandas as pd
|
||||||
|
import lancedb
|
||||||
|
from lancedb.pydantic import LanceModel, Vector
|
||||||
|
from lancedb.embeddings import get_registry
|
||||||
|
|
||||||
|
db = lancedb.connect("/tmp/db")
|
||||||
|
model = get_registry().get("sentence-transformers").create(name="BAAI/bge-small-en-v1.5", device="cpu")
|
||||||
|
|
||||||
|
class Docs(LanceModel):
|
||||||
|
text: str = model.SourceField()
|
||||||
|
vector: Vector(model.ndims()) = model.VectorField()
|
||||||
|
|
||||||
|
table = db.create_table("docs", schema=Docs)
|
||||||
|
|
||||||
|
# considering chunks are in list format
|
||||||
|
df = pd.DataFrame({'text':chunks})
|
||||||
|
table.add(data=df)
|
||||||
|
|
||||||
|
query = "What is issue date of lease?"
|
||||||
|
actual = table.search(query).limit(1).to_list()[0]
|
||||||
|
print(actual.text)
|
||||||
|
```
|
||||||
|
|
||||||
|
Check Colab for the complete code
|
||||||
|
|
||||||
|
[](https://colab.research.google.com/github/lancedb/vectordb-recipes/blob/main/tutorials/RAG-from-Scratch/RAG_from_Scratch.ipynb)
|
||||||
@@ -20,7 +20,11 @@ excluded_globs = [
|
|||||||
"../src/reranking/*.md",
|
"../src/reranking/*.md",
|
||||||
"../src/guides/tuning_retrievers/*.md",
|
"../src/guides/tuning_retrievers/*.md",
|
||||||
"../src/embeddings/available_embedding_models/text_embedding_functions/*.md",
|
"../src/embeddings/available_embedding_models/text_embedding_functions/*.md",
|
||||||
"../src/embeddings/available_embedding_models/multimodal_embedding_functions/*.md"
|
"../src/embeddings/available_embedding_models/multimodal_embedding_functions/*.md",
|
||||||
|
"../src/rag/*.md",
|
||||||
|
"../src/rag/advanced_techniques/*.md"
|
||||||
|
|
||||||
|
|
||||||
]
|
]
|
||||||
|
|
||||||
python_prefix = "py"
|
python_prefix = "py"
|
||||||
|
|||||||
@@ -2,7 +2,7 @@
|
|||||||
name = "lancedb-jni"
|
name = "lancedb-jni"
|
||||||
description = "JNI bindings for LanceDB"
|
description = "JNI bindings for LanceDB"
|
||||||
# TODO modify lancedb/Cargo.toml for version and dependencies
|
# TODO modify lancedb/Cargo.toml for version and dependencies
|
||||||
version = "0.4.18"
|
version = "0.10.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
repository.workspace = true
|
repository.workspace = true
|
||||||
readme.workspace = true
|
readme.workspace = true
|
||||||
|
|||||||
@@ -8,7 +8,7 @@
|
|||||||
<parent>
|
<parent>
|
||||||
<groupId>com.lancedb</groupId>
|
<groupId>com.lancedb</groupId>
|
||||||
<artifactId>lancedb-parent</artifactId>
|
<artifactId>lancedb-parent</artifactId>
|
||||||
<version>0.0.3</version>
|
<version>0.10.0</version>
|
||||||
<relativePath>../pom.xml</relativePath>
|
<relativePath>../pom.xml</relativePath>
|
||||||
</parent>
|
</parent>
|
||||||
|
|
||||||
@@ -44,7 +44,7 @@
|
|||||||
<dependency>
|
<dependency>
|
||||||
<groupId>org.junit.jupiter</groupId>
|
<groupId>org.junit.jupiter</groupId>
|
||||||
<artifactId>junit-jupiter</artifactId>
|
<artifactId>junit-jupiter</artifactId>
|
||||||
<scope>test</scope>
|
<scope>test</scope>
|
||||||
</dependency>
|
</dependency>
|
||||||
</dependencies>
|
</dependencies>
|
||||||
|
|
||||||
|
|||||||
12
java/pom.xml
12
java/pom.xml
@@ -6,7 +6,7 @@
|
|||||||
|
|
||||||
<groupId>com.lancedb</groupId>
|
<groupId>com.lancedb</groupId>
|
||||||
<artifactId>lancedb-parent</artifactId>
|
<artifactId>lancedb-parent</artifactId>
|
||||||
<version>0.0.3</version>
|
<version>0.10.0</version>
|
||||||
<packaging>pom</packaging>
|
<packaging>pom</packaging>
|
||||||
|
|
||||||
<name>LanceDB Parent</name>
|
<name>LanceDB Parent</name>
|
||||||
@@ -92,7 +92,7 @@
|
|||||||
</repository>
|
</repository>
|
||||||
</distributionManagement>
|
</distributionManagement>
|
||||||
|
|
||||||
<build>
|
<build>
|
||||||
<plugins>
|
<plugins>
|
||||||
<plugin>
|
<plugin>
|
||||||
<groupId>org.apache.maven.plugins</groupId>
|
<groupId>org.apache.maven.plugins</groupId>
|
||||||
@@ -167,7 +167,8 @@
|
|||||||
<version>3.2.5</version>
|
<version>3.2.5</version>
|
||||||
<configuration>
|
<configuration>
|
||||||
<argLine>--add-opens=java.base/java.nio=ALL-UNNAMED</argLine>
|
<argLine>--add-opens=java.base/java.nio=ALL-UNNAMED</argLine>
|
||||||
<forkNode implementation="org.apache.maven.plugin.surefire.extensions.SurefireForkNodeFactory"/>
|
<forkNode
|
||||||
|
implementation="org.apache.maven.plugin.surefire.extensions.SurefireForkNodeFactory" />
|
||||||
<useSystemClassLoader>false</useSystemClassLoader>
|
<useSystemClassLoader>false</useSystemClassLoader>
|
||||||
</configuration>
|
</configuration>
|
||||||
</plugin>
|
</plugin>
|
||||||
@@ -183,7 +184,7 @@
|
|||||||
</pluginManagement>
|
</pluginManagement>
|
||||||
</build>
|
</build>
|
||||||
|
|
||||||
<profiles>
|
<profiles>
|
||||||
<profile>
|
<profile>
|
||||||
<id>jdk8</id>
|
<id>jdk8</id>
|
||||||
<activation>
|
<activation>
|
||||||
@@ -210,7 +211,8 @@
|
|||||||
<version>3.2.5</version>
|
<version>3.2.5</version>
|
||||||
<configuration>
|
<configuration>
|
||||||
<argLine>--add-opens=java.base/java.nio=ALL-UNNAMED</argLine>
|
<argLine>--add-opens=java.base/java.nio=ALL-UNNAMED</argLine>
|
||||||
<forkNode implementation="org.apache.maven.plugin.surefire.extensions.SurefireForkNodeFactory" />
|
<forkNode
|
||||||
|
implementation="org.apache.maven.plugin.surefire.extensions.SurefireForkNodeFactory" />
|
||||||
<useSystemClassLoader>false</useSystemClassLoader>
|
<useSystemClassLoader>false</useSystemClassLoader>
|
||||||
</configuration>
|
</configuration>
|
||||||
</plugin>
|
</plugin>
|
||||||
|
|||||||
4
node/package-lock.json
generated
4
node/package-lock.json
generated
@@ -1,12 +1,12 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"lockfileVersion": 3,
|
"lockfileVersion": 3,
|
||||||
"requires": true,
|
"requires": true,
|
||||||
"packages": {
|
"packages": {
|
||||||
"": {
|
"": {
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"cpu": [
|
"cpu": [
|
||||||
"x64",
|
"x64",
|
||||||
"arm64"
|
"arm64"
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "vectordb",
|
"name": "vectordb",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"description": " Serverless, low-latency vector database for AI applications",
|
"description": " Serverless, low-latency vector database for AI applications",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"types": "dist/index.d.ts",
|
"types": "dist/index.d.ts",
|
||||||
|
|||||||
@@ -60,7 +60,7 @@ export {
|
|||||||
type MakeArrowTableOptions
|
type MakeArrowTableOptions
|
||||||
} from "./arrow";
|
} from "./arrow";
|
||||||
|
|
||||||
const defaultAwsRegion = "us-west-2";
|
const defaultAwsRegion = "us-east-1";
|
||||||
|
|
||||||
const defaultRequestTimeout = 10_000
|
const defaultRequestTimeout = 10_000
|
||||||
|
|
||||||
@@ -111,7 +111,7 @@ export interface ConnectionOptions {
|
|||||||
*/
|
*/
|
||||||
apiKey?: string
|
apiKey?: string
|
||||||
|
|
||||||
/** Region to connect */
|
/** Region to connect. Default is 'us-east-1' */
|
||||||
region?: string
|
region?: string
|
||||||
|
|
||||||
/**
|
/**
|
||||||
@@ -197,28 +197,31 @@ export async function connect(
|
|||||||
export async function connect(
|
export async function connect(
|
||||||
arg: string | Partial<ConnectionOptions>
|
arg: string | Partial<ConnectionOptions>
|
||||||
): Promise<Connection> {
|
): Promise<Connection> {
|
||||||
let opts: ConnectionOptions;
|
let partOpts: Partial<ConnectionOptions>;
|
||||||
if (typeof arg === "string") {
|
if (typeof arg === "string") {
|
||||||
opts = { uri: arg };
|
partOpts = { uri: arg };
|
||||||
} else {
|
} else {
|
||||||
const keys = Object.keys(arg);
|
const keys = Object.keys(arg);
|
||||||
if (keys.length === 1 && keys[0] === "uri" && typeof arg.uri === "string") {
|
if (keys.length === 1 && keys[0] === "uri" && typeof arg.uri === "string") {
|
||||||
opts = { uri: arg.uri };
|
partOpts = { uri: arg.uri };
|
||||||
} else {
|
} else {
|
||||||
opts = Object.assign(
|
partOpts = arg;
|
||||||
{
|
|
||||||
uri: "",
|
|
||||||
awsCredentials: undefined,
|
|
||||||
awsRegion: defaultAwsRegion,
|
|
||||||
apiKey: undefined,
|
|
||||||
region: defaultAwsRegion,
|
|
||||||
timeout: defaultRequestTimeout
|
|
||||||
},
|
|
||||||
arg
|
|
||||||
);
|
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
let defaultRegion = process.env.AWS_REGION ?? process.env.AWS_DEFAULT_REGION;
|
||||||
|
defaultRegion = (defaultRegion ?? "").trim() !== "" ? defaultRegion : defaultAwsRegion;
|
||||||
|
|
||||||
|
const opts: ConnectionOptions = {
|
||||||
|
uri: partOpts.uri ?? "",
|
||||||
|
awsCredentials: partOpts.awsCredentials ?? undefined,
|
||||||
|
awsRegion: partOpts.awsRegion ?? defaultRegion,
|
||||||
|
apiKey: partOpts.apiKey ?? undefined,
|
||||||
|
region: partOpts.region ?? defaultRegion,
|
||||||
|
timeout: partOpts.timeout ?? defaultRequestTimeout,
|
||||||
|
readConsistencyInterval: partOpts.readConsistencyInterval ?? undefined,
|
||||||
|
storageOptions: partOpts.storageOptions ?? undefined
|
||||||
|
}
|
||||||
if (opts.uri.startsWith("db://")) {
|
if (opts.uri.startsWith("db://")) {
|
||||||
// Remote connection
|
// Remote connection
|
||||||
return new RemoteConnection(opts);
|
return new RemoteConnection(opts);
|
||||||
|
|||||||
@@ -82,7 +82,7 @@ async function callWithMiddlewares (
|
|||||||
|
|
||||||
interface MiddlewareInvocationOptions {
|
interface MiddlewareInvocationOptions {
|
||||||
responseType?: ResponseType
|
responseType?: ResponseType
|
||||||
timeout?: number,
|
timeout?: number
|
||||||
}
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
@@ -130,7 +130,7 @@ export class HttpLancedbClient {
|
|||||||
url: string,
|
url: string,
|
||||||
apiKey: string,
|
apiKey: string,
|
||||||
timeout?: number,
|
timeout?: number,
|
||||||
private readonly _dbName?: string,
|
private readonly _dbName?: string
|
||||||
|
|
||||||
) {
|
) {
|
||||||
this._url = url
|
this._url = url
|
||||||
@@ -237,7 +237,7 @@ export class HttpLancedbClient {
|
|||||||
try {
|
try {
|
||||||
response = await callWithMiddlewares(req, this._middlewares, {
|
response = await callWithMiddlewares(req, this._middlewares, {
|
||||||
responseType,
|
responseType,
|
||||||
timeout: this._timeout,
|
timeout: this._timeout
|
||||||
})
|
})
|
||||||
|
|
||||||
// return response
|
// return response
|
||||||
|
|||||||
@@ -112,8 +112,8 @@ describe("LanceDB client", function () {
|
|||||||
name: 'name_2',
|
name: 'name_2',
|
||||||
price: 10,
|
price: 10,
|
||||||
is_active: true,
|
is_active: true,
|
||||||
vector: [ 0, 0.1 ]
|
vector: [0, 0.1]
|
||||||
},
|
}
|
||||||
]);
|
]);
|
||||||
assert.equal(await table2.countRows(), 3);
|
assert.equal(await table2.countRows(), 3);
|
||||||
});
|
});
|
||||||
|
|||||||
@@ -107,7 +107,7 @@ describe("given a connection", () => {
|
|||||||
const data = [...Array(10000).keys()].map((i) => ({ id: i }));
|
const data = [...Array(10000).keys()].map((i) => ({ id: i }));
|
||||||
|
|
||||||
// Create in v1 mode
|
// Create in v1 mode
|
||||||
let table = await db.createTable("test", data);
|
let table = await db.createTable("test", data, { useLegacyFormat: true });
|
||||||
|
|
||||||
const isV2 = async (table: Table) => {
|
const isV2 = async (table: Table) => {
|
||||||
const data = await table.query().toArrow({ maxBatchLength: 100000 });
|
const data = await table.query().toArrow({ maxBatchLength: 100000 });
|
||||||
@@ -118,7 +118,7 @@ describe("given a connection", () => {
|
|||||||
await expect(isV2(table)).resolves.toBe(false);
|
await expect(isV2(table)).resolves.toBe(false);
|
||||||
|
|
||||||
// Create in v2 mode
|
// Create in v2 mode
|
||||||
table = await db.createTable("test_v2", data, { useLegacyFormat: false });
|
table = await db.createTable("test_v2", data);
|
||||||
|
|
||||||
await expect(isV2(table)).resolves.toBe(true);
|
await expect(isV2(table)).resolves.toBe(true);
|
||||||
|
|
||||||
|
|||||||
@@ -872,7 +872,7 @@ describe.each([arrow13, arrow14, arrow15, arrow16, arrow17])(
|
|||||||
];
|
];
|
||||||
const table = await db.createTable("test", data);
|
const table = await db.createTable("test", data);
|
||||||
await table.createIndex("text", {
|
await table.createIndex("text", {
|
||||||
config: Index.fts({ withPositions: false }),
|
config: Index.fts({ withPosition: false }),
|
||||||
});
|
});
|
||||||
|
|
||||||
const results = await table.search("hello").toArray();
|
const results = await table.search("hello").toArray();
|
||||||
|
|||||||
@@ -44,11 +44,12 @@ export interface CreateTableOptions {
|
|||||||
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
* The available options are described at https://lancedb.github.io/lancedb/guides/storage/
|
||||||
*/
|
*/
|
||||||
storageOptions?: Record<string, string>;
|
storageOptions?: Record<string, string>;
|
||||||
|
|
||||||
/**
|
/**
|
||||||
* The version of the data storage format to use.
|
* The version of the data storage format to use.
|
||||||
*
|
*
|
||||||
* The default is `legacy`, which is Lance format v1.
|
* The default is `stable`.
|
||||||
* `stable` is the new format, which is Lance format v2.
|
* Set to "legacy" to use the old format.
|
||||||
*/
|
*/
|
||||||
dataStorageVersion?: string;
|
dataStorageVersion?: string;
|
||||||
|
|
||||||
@@ -64,9 +65,9 @@ export interface CreateTableOptions {
|
|||||||
/**
|
/**
|
||||||
* If true then data files will be written with the legacy format
|
* If true then data files will be written with the legacy format
|
||||||
*
|
*
|
||||||
* The default is true while the new format is in beta
|
* The default is false.
|
||||||
*
|
*
|
||||||
* Deprecated.
|
* Deprecated. Use data storage version instead.
|
||||||
*/
|
*/
|
||||||
useLegacyFormat?: boolean;
|
useLegacyFormat?: boolean;
|
||||||
schema?: SchemaLike;
|
schema?: SchemaLike;
|
||||||
@@ -266,7 +267,7 @@ export class LocalConnection extends Connection {
|
|||||||
throw new Error("data is required");
|
throw new Error("data is required");
|
||||||
}
|
}
|
||||||
const { buf, mode } = await Table.parseTableData(data, options);
|
const { buf, mode } = await Table.parseTableData(data, options);
|
||||||
let dataStorageVersion = "legacy";
|
let dataStorageVersion = "stable";
|
||||||
if (options?.dataStorageVersion !== undefined) {
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
dataStorageVersion = options.dataStorageVersion;
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
} else if (options?.useLegacyFormat !== undefined) {
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
@@ -303,7 +304,7 @@ export class LocalConnection extends Connection {
|
|||||||
metadata = registry.getTableMetadata([embeddingFunction]);
|
metadata = registry.getTableMetadata([embeddingFunction]);
|
||||||
}
|
}
|
||||||
|
|
||||||
let dataStorageVersion = "legacy";
|
let dataStorageVersion = "stable";
|
||||||
if (options?.dataStorageVersion !== undefined) {
|
if (options?.dataStorageVersion !== undefined) {
|
||||||
dataStorageVersion = options.dataStorageVersion;
|
dataStorageVersion = options.dataStorageVersion;
|
||||||
} else if (options?.useLegacyFormat !== undefined) {
|
} else if (options?.useLegacyFormat !== undefined) {
|
||||||
|
|||||||
@@ -113,22 +113,218 @@ export interface IvfPqOptions {
|
|||||||
sampleRate?: number;
|
sampleRate?: number;
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Options to create an `HNSW_PQ` index
|
||||||
|
*/
|
||||||
export interface HnswPqOptions {
|
export interface HnswPqOptions {
|
||||||
|
/**
|
||||||
|
* The distance metric used to train the index.
|
||||||
|
*
|
||||||
|
* Default value is "l2".
|
||||||
|
*
|
||||||
|
* The following distance types are available:
|
||||||
|
*
|
||||||
|
* "l2" - Euclidean distance. This is a very common distance metric that
|
||||||
|
* accounts for both magnitude and direction when determining the distance
|
||||||
|
* between vectors. L2 distance has a range of [0, ∞).
|
||||||
|
*
|
||||||
|
* "cosine" - Cosine distance. Cosine distance is a distance metric
|
||||||
|
* calculated from the cosine similarity between two vectors. Cosine
|
||||||
|
* similarity is a measure of similarity between two non-zero vectors of an
|
||||||
|
* inner product space. It is defined to equal the cosine of the angle
|
||||||
|
* between them. Unlike L2, the cosine distance is not affected by the
|
||||||
|
* magnitude of the vectors. Cosine distance has a range of [0, 2].
|
||||||
|
*
|
||||||
|
* "dot" - Dot product. Dot distance is the dot product of two vectors. Dot
|
||||||
|
* distance has a range of (-∞, ∞). If the vectors are normalized (i.e. their
|
||||||
|
* L2 norm is 1), then dot distance is equivalent to the cosine distance.
|
||||||
|
*/
|
||||||
distanceType?: "l2" | "cosine" | "dot";
|
distanceType?: "l2" | "cosine" | "dot";
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of IVF partitions to create.
|
||||||
|
*
|
||||||
|
* For HNSW, we recommend a small number of partitions. Setting this to 1 works
|
||||||
|
* well for most tables. For very large tables, training just one HNSW graph
|
||||||
|
* will require too much memory. Each partition becomes its own HNSW graph, so
|
||||||
|
* setting this value higher reduces the peak memory use of training.
|
||||||
|
*
|
||||||
|
*/
|
||||||
numPartitions?: number;
|
numPartitions?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Number of sub-vectors of PQ.
|
||||||
|
*
|
||||||
|
* This value controls how much the vector is compressed during the quantization step.
|
||||||
|
* The more sub vectors there are the less the vector is compressed. The default is
|
||||||
|
* the dimension of the vector divided by 16. If the dimension is not evenly divisible
|
||||||
|
* by 16 we use the dimension divded by 8.
|
||||||
|
*
|
||||||
|
* The above two cases are highly preferred. Having 8 or 16 values per subvector allows
|
||||||
|
* us to use efficient SIMD instructions.
|
||||||
|
*
|
||||||
|
* If the dimension is not visible by 8 then we use 1 subvector. This is not ideal and
|
||||||
|
* will likely result in poor performance.
|
||||||
|
*
|
||||||
|
*/
|
||||||
numSubVectors?: number;
|
numSubVectors?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Max iterations to train kmeans.
|
||||||
|
*
|
||||||
|
* The default value is 50.
|
||||||
|
*
|
||||||
|
* When training an IVF index we use kmeans to calculate the partitions. This parameter
|
||||||
|
* controls how many iterations of kmeans to run.
|
||||||
|
*
|
||||||
|
* Increasing this might improve the quality of the index but in most cases the parameter
|
||||||
|
* is unused because kmeans will converge with fewer iterations. The parameter is only
|
||||||
|
* used in cases where kmeans does not appear to converge. In those cases it is unlikely
|
||||||
|
* that setting this larger will lead to the index converging anyways.
|
||||||
|
*
|
||||||
|
*/
|
||||||
maxIterations?: number;
|
maxIterations?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The rate used to calculate the number of training vectors for kmeans.
|
||||||
|
*
|
||||||
|
* Default value is 256.
|
||||||
|
*
|
||||||
|
* When an IVF index is trained, we need to calculate partitions. These are groups
|
||||||
|
* of vectors that are similar to each other. To do this we use an algorithm called kmeans.
|
||||||
|
*
|
||||||
|
* Running kmeans on a large dataset can be slow. To speed this up we run kmeans on a
|
||||||
|
* random sample of the data. This parameter controls the size of the sample. The total
|
||||||
|
* number of vectors used to train the index is `sample_rate * num_partitions`.
|
||||||
|
*
|
||||||
|
* Increasing this value might improve the quality of the index but in most cases the
|
||||||
|
* default should be sufficient.
|
||||||
|
*
|
||||||
|
*/
|
||||||
sampleRate?: number;
|
sampleRate?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of neighbors to select for each vector in the HNSW graph.
|
||||||
|
*
|
||||||
|
* The default value is 20.
|
||||||
|
*
|
||||||
|
* This value controls the tradeoff between search speed and accuracy.
|
||||||
|
* The higher the value the more accurate the search but the slower it will be.
|
||||||
|
*
|
||||||
|
*/
|
||||||
m?: number;
|
m?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of candidates to evaluate during the construction of the HNSW graph.
|
||||||
|
*
|
||||||
|
* The default value is 300.
|
||||||
|
*
|
||||||
|
* This value controls the tradeoff between build speed and accuracy.
|
||||||
|
* The higher the value the more accurate the build but the slower it will be.
|
||||||
|
* 150 to 300 is the typical range. 100 is a minimum for good quality search
|
||||||
|
* results. In most cases, there is no benefit to setting this higher than 500.
|
||||||
|
* This value should be set to a value that is not less than `ef` in the search phase.
|
||||||
|
*
|
||||||
|
*/
|
||||||
efConstruction?: number;
|
efConstruction?: number;
|
||||||
}
|
}
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Options to create an `HNSW_SQ` index
|
||||||
|
*/
|
||||||
export interface HnswSqOptions {
|
export interface HnswSqOptions {
|
||||||
|
/**
|
||||||
|
* The distance metric used to train the index.
|
||||||
|
*
|
||||||
|
* Default value is "l2".
|
||||||
|
*
|
||||||
|
* The following distance types are available:
|
||||||
|
*
|
||||||
|
* "l2" - Euclidean distance. This is a very common distance metric that
|
||||||
|
* accounts for both magnitude and direction when determining the distance
|
||||||
|
* between vectors. L2 distance has a range of [0, ∞).
|
||||||
|
*
|
||||||
|
* "cosine" - Cosine distance. Cosine distance is a distance metric
|
||||||
|
* calculated from the cosine similarity between two vectors. Cosine
|
||||||
|
* similarity is a measure of similarity between two non-zero vectors of an
|
||||||
|
* inner product space. It is defined to equal the cosine of the angle
|
||||||
|
* between them. Unlike L2, the cosine distance is not affected by the
|
||||||
|
* magnitude of the vectors. Cosine distance has a range of [0, 2].
|
||||||
|
*
|
||||||
|
* "dot" - Dot product. Dot distance is the dot product of two vectors. Dot
|
||||||
|
* distance has a range of (-∞, ∞). If the vectors are normalized (i.e. their
|
||||||
|
* L2 norm is 1), then dot distance is equivalent to the cosine distance.
|
||||||
|
*/
|
||||||
distanceType?: "l2" | "cosine" | "dot";
|
distanceType?: "l2" | "cosine" | "dot";
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of IVF partitions to create.
|
||||||
|
*
|
||||||
|
* For HNSW, we recommend a small number of partitions. Setting this to 1 works
|
||||||
|
* well for most tables. For very large tables, training just one HNSW graph
|
||||||
|
* will require too much memory. Each partition becomes its own HNSW graph, so
|
||||||
|
* setting this value higher reduces the peak memory use of training.
|
||||||
|
*
|
||||||
|
*/
|
||||||
numPartitions?: number;
|
numPartitions?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* Max iterations to train kmeans.
|
||||||
|
*
|
||||||
|
* The default value is 50.
|
||||||
|
*
|
||||||
|
* When training an IVF index we use kmeans to calculate the partitions. This parameter
|
||||||
|
* controls how many iterations of kmeans to run.
|
||||||
|
*
|
||||||
|
* Increasing this might improve the quality of the index but in most cases the parameter
|
||||||
|
* is unused because kmeans will converge with fewer iterations. The parameter is only
|
||||||
|
* used in cases where kmeans does not appear to converge. In those cases it is unlikely
|
||||||
|
* that setting this larger will lead to the index converging anyways.
|
||||||
|
*
|
||||||
|
*/
|
||||||
maxIterations?: number;
|
maxIterations?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The rate used to calculate the number of training vectors for kmeans.
|
||||||
|
*
|
||||||
|
* Default value is 256.
|
||||||
|
*
|
||||||
|
* When an IVF index is trained, we need to calculate partitions. These are groups
|
||||||
|
* of vectors that are similar to each other. To do this we use an algorithm called kmeans.
|
||||||
|
*
|
||||||
|
* Running kmeans on a large dataset can be slow. To speed this up we run kmeans on a
|
||||||
|
* random sample of the data. This parameter controls the size of the sample. The total
|
||||||
|
* number of vectors used to train the index is `sample_rate * num_partitions`.
|
||||||
|
*
|
||||||
|
* Increasing this value might improve the quality of the index but in most cases the
|
||||||
|
* default should be sufficient.
|
||||||
|
*
|
||||||
|
*/
|
||||||
sampleRate?: number;
|
sampleRate?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of neighbors to select for each vector in the HNSW graph.
|
||||||
|
*
|
||||||
|
* The default value is 20.
|
||||||
|
*
|
||||||
|
* This value controls the tradeoff between search speed and accuracy.
|
||||||
|
* The higher the value the more accurate the search but the slower it will be.
|
||||||
|
*
|
||||||
|
*/
|
||||||
m?: number;
|
m?: number;
|
||||||
|
|
||||||
|
/**
|
||||||
|
* The number of candidates to evaluate during the construction of the HNSW graph.
|
||||||
|
*
|
||||||
|
* The default value is 300.
|
||||||
|
*
|
||||||
|
* This value controls the tradeoff between build speed and accuracy.
|
||||||
|
* The higher the value the more accurate the build but the slower it will be.
|
||||||
|
* 150 to 300 is the typical range. 100 is a minimum for good quality search
|
||||||
|
* results. In most cases, there is no benefit to setting this higher than 500.
|
||||||
|
* This value should be set to a value that is not less than `ef` in the search phase.
|
||||||
|
*
|
||||||
|
*/
|
||||||
efConstruction?: number;
|
efConstruction?: number;
|
||||||
}
|
}
|
||||||
|
|
||||||
@@ -142,7 +338,7 @@ export interface FtsOptions {
|
|||||||
* If set to false, the index will not store the positions of the tokens in the text,
|
* If set to false, the index will not store the positions of the tokens in the text,
|
||||||
* which will make the index smaller and faster to build, but will not support phrase queries.
|
* which will make the index smaller and faster to build, but will not support phrase queries.
|
||||||
*/
|
*/
|
||||||
withPositions?: boolean;
|
withPosition?: boolean;
|
||||||
}
|
}
|
||||||
|
|
||||||
export class Index {
|
export class Index {
|
||||||
@@ -244,12 +440,16 @@ export class Index {
|
|||||||
* For now, the full text search index only supports English, and doesn't support phrase search.
|
* For now, the full text search index only supports English, and doesn't support phrase search.
|
||||||
*/
|
*/
|
||||||
static fts(options?: Partial<FtsOptions>) {
|
static fts(options?: Partial<FtsOptions>) {
|
||||||
return new Index(LanceDbIndex.fts(options?.withPositions));
|
return new Index(LanceDbIndex.fts(options?.withPosition));
|
||||||
}
|
}
|
||||||
|
|
||||||
/**
|
/**
|
||||||
*
|
*
|
||||||
* Create a hnswpq index
|
* Create a hnswPq index
|
||||||
|
*
|
||||||
|
* HNSW-PQ stands for Hierarchical Navigable Small World - Product Quantization.
|
||||||
|
* It is a variant of the HNSW algorithm that uses product quantization to compress
|
||||||
|
* the vectors.
|
||||||
*
|
*
|
||||||
*/
|
*/
|
||||||
static hnswPq(options?: Partial<HnswPqOptions>) {
|
static hnswPq(options?: Partial<HnswPqOptions>) {
|
||||||
@@ -268,7 +468,11 @@ export class Index {
|
|||||||
|
|
||||||
/**
|
/**
|
||||||
*
|
*
|
||||||
* Create a hnswsq index
|
* Create a hnswSq index
|
||||||
|
*
|
||||||
|
* HNSW-SQ stands for Hierarchical Navigable Small World - Scalar Quantization.
|
||||||
|
* It is a variant of the HNSW algorithm that uses scalar quantization to compress
|
||||||
|
* the vectors.
|
||||||
*
|
*
|
||||||
*/
|
*/
|
||||||
static hnswSq(options?: Partial<HnswSqOptions>) {
|
static hnswSq(options?: Partial<HnswSqOptions>) {
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-arm64",
|
"name": "@lancedb/lancedb-darwin-arm64",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.darwin-arm64.node",
|
"main": "lancedb.darwin-arm64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-darwin-x64",
|
"name": "@lancedb/lancedb-darwin-x64",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"os": ["darwin"],
|
"os": ["darwin"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.darwin-x64.node",
|
"main": "lancedb.darwin-x64.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
"name": "@lancedb/lancedb-linux-arm64-gnu",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["arm64"],
|
"cpu": ["arm64"],
|
||||||
"main": "lancedb.linux-arm64-gnu.node",
|
"main": "lancedb.linux-arm64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-linux-x64-gnu",
|
"name": "@lancedb/lancedb-linux-x64-gnu",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"os": ["linux"],
|
"os": ["linux"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.linux-x64-gnu.node",
|
"main": "lancedb.linux-x64-gnu.node",
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
{
|
{
|
||||||
"name": "@lancedb/lancedb-win32-x64-msvc",
|
"name": "@lancedb/lancedb-win32-x64-msvc",
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"os": ["win32"],
|
"os": ["win32"],
|
||||||
"cpu": ["x64"],
|
"cpu": ["x64"],
|
||||||
"main": "lancedb.win32-x64-msvc.node",
|
"main": "lancedb.win32-x64-msvc.node",
|
||||||
|
|||||||
@@ -10,7 +10,7 @@
|
|||||||
"vector database",
|
"vector database",
|
||||||
"ann"
|
"ann"
|
||||||
],
|
],
|
||||||
"version": "0.10.0-beta.1",
|
"version": "0.10.0",
|
||||||
"main": "dist/index.js",
|
"main": "dist/index.js",
|
||||||
"exports": {
|
"exports": {
|
||||||
".": "./dist/index.js",
|
".": "./dist/index.js",
|
||||||
|
|||||||
@@ -130,6 +130,7 @@ impl Connection {
|
|||||||
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
.map_err(|e| napi::Error::from_reason(format!("Failed to read IPC file: {}", e)))?;
|
||||||
let mode = Self::parse_create_mode_str(&mode)?;
|
let mode = Self::parse_create_mode_str(&mode)?;
|
||||||
let mut builder = self.get_inner()?.create_table(&name, batches).mode(mode);
|
let mut builder = self.get_inner()?.create_table(&name, batches).mode(mode);
|
||||||
|
|
||||||
if let Some(storage_options) = storage_options {
|
if let Some(storage_options) = storage_options {
|
||||||
for (key, value) in storage_options {
|
for (key, value) in storage_options {
|
||||||
builder = builder.storage_option(key, value);
|
builder = builder.storage_option(key, value);
|
||||||
|
|||||||
@@ -156,7 +156,7 @@ impl Table {
|
|||||||
&self,
|
&self,
|
||||||
only_if: Option<String>,
|
only_if: Option<String>,
|
||||||
columns: Vec<(String, String)>,
|
columns: Vec<(String, String)>,
|
||||||
) -> napi::Result<()> {
|
) -> napi::Result<u64> {
|
||||||
let mut op = self.inner_ref()?.update();
|
let mut op = self.inner_ref()?.update();
|
||||||
if let Some(only_if) = only_if {
|
if let Some(only_if) = only_if {
|
||||||
op = op.only_if(only_if);
|
op = op.only_if(only_if);
|
||||||
|
|||||||
@@ -1,5 +1,5 @@
|
|||||||
[tool.bumpversion]
|
[tool.bumpversion]
|
||||||
current_version = "0.13.0"
|
current_version = "0.14.0-beta.0"
|
||||||
parse = """(?x)
|
parse = """(?x)
|
||||||
(?P<major>0|[1-9]\\d*)\\.
|
(?P<major>0|[1-9]\\d*)\\.
|
||||||
(?P<minor>0|[1-9]\\d*)\\.
|
(?P<minor>0|[1-9]\\d*)\\.
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb-python"
|
name = "lancedb-python"
|
||||||
version = "0.13.0"
|
version = "0.14.0-beta.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
description = "Python bindings for LanceDB"
|
description = "Python bindings for LanceDB"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
|
|||||||
@@ -3,7 +3,7 @@ name = "lancedb"
|
|||||||
# version in Cargo.toml
|
# version in Cargo.toml
|
||||||
dependencies = [
|
dependencies = [
|
||||||
"deprecation",
|
"deprecation",
|
||||||
"pylance==0.17.0",
|
"pylance==0.18.0",
|
||||||
"requests>=2.31.0",
|
"requests>=2.31.0",
|
||||||
"retry>=0.9.2",
|
"retry>=0.9.2",
|
||||||
"tqdm>=4.27.0",
|
"tqdm>=4.27.0",
|
||||||
|
|||||||
@@ -610,14 +610,13 @@ class AsyncConnection(object):
|
|||||||
connection will be inherited by the table, but can be overridden here.
|
connection will be inherited by the table, but can be overridden here.
|
||||||
See available options at
|
See available options at
|
||||||
https://lancedb.github.io/lancedb/guides/storage/
|
https://lancedb.github.io/lancedb/guides/storage/
|
||||||
data_storage_version: optional, str, default "legacy"
|
data_storage_version: optional, str, default "stable"
|
||||||
The version of the data storage format to use. Newer versions are more
|
The version of the data storage format to use. Newer versions are more
|
||||||
efficient but require newer versions of lance to read. The default is
|
efficient but require newer versions of lance to read. The default is
|
||||||
"legacy" which will use the legacy v1 version. See the user guide
|
"stable" which will use the legacy v2 version. See the user guide
|
||||||
for more details.
|
for more details.
|
||||||
use_legacy_format: bool, optional, default True. (Deprecated)
|
use_legacy_format: bool, optional, default False. (Deprecated)
|
||||||
If True, use the legacy format for the table. If False, use the new format.
|
If True, use the legacy format for the table. If False, use the new format.
|
||||||
The default is True while the new format is in beta.
|
|
||||||
This method is deprecated, use `data_storage_version` instead.
|
This method is deprecated, use `data_storage_version` instead.
|
||||||
enable_v2_manifest_paths: bool, optional, default False
|
enable_v2_manifest_paths: bool, optional, default False
|
||||||
Use the new V2 manifest paths. These paths provide more efficient
|
Use the new V2 manifest paths. These paths provide more efficient
|
||||||
@@ -759,9 +758,7 @@ class AsyncConnection(object):
|
|||||||
mode = "exist_ok"
|
mode = "exist_ok"
|
||||||
|
|
||||||
if not data_storage_version:
|
if not data_storage_version:
|
||||||
data_storage_version = (
|
data_storage_version = "legacy" if use_legacy_format else "stable"
|
||||||
"legacy" if use_legacy_format is None or use_legacy_format else "stable"
|
|
||||||
)
|
|
||||||
|
|
||||||
if data is None:
|
if data is None:
|
||||||
new_table = await self._inner.create_empty_table(
|
new_table = await self._inner.create_empty_table(
|
||||||
|
|||||||
259
python/python/lancedb/dependencies.py
Normal file
259
python/python/lancedb/dependencies.py
Normal file
@@ -0,0 +1,259 @@
|
|||||||
|
# SPDX-License-Identifier: Apache-2.0
|
||||||
|
# SPDX-FileCopyrightText: Copyright The Lance Authors
|
||||||
|
#
|
||||||
|
# The following code is originally from https://github.com/pola-rs/polars/blob/ea4389c31b0e87ddf20a85e4c3797b285966edb6/py-polars/polars/dependencies.py
|
||||||
|
# and is licensed under the MIT license:
|
||||||
|
#
|
||||||
|
# License: MIT, Copyright (c) 2020 Ritchie Vink
|
||||||
|
# https://github.com/pola-rs/polars/blob/main/LICENSE
|
||||||
|
#
|
||||||
|
# It has been modified by the LanceDB developers
|
||||||
|
# to fit the needs of the LanceDB project.
|
||||||
|
|
||||||
|
|
||||||
|
from __future__ import annotations
|
||||||
|
|
||||||
|
import re
|
||||||
|
import sys
|
||||||
|
from functools import lru_cache
|
||||||
|
from importlib import import_module
|
||||||
|
from importlib.util import find_spec
|
||||||
|
from types import ModuleType
|
||||||
|
from typing import TYPE_CHECKING, Any, ClassVar, Hashable, cast
|
||||||
|
|
||||||
|
_NUMPY_AVAILABLE = True
|
||||||
|
_PANDAS_AVAILABLE = True
|
||||||
|
_POLARS_AVAILABLE = True
|
||||||
|
_TORCH_AVAILABLE = True
|
||||||
|
_HUGGING_FACE_AVAILABLE = True
|
||||||
|
_TENSORFLOW_AVAILABLE = True
|
||||||
|
_RAY_AVAILABLE = True
|
||||||
|
|
||||||
|
|
||||||
|
class _LazyModule(ModuleType):
|
||||||
|
"""
|
||||||
|
Module that can act both as a lazy-loader and as a proxy.
|
||||||
|
|
||||||
|
Notes
|
||||||
|
-----
|
||||||
|
We do NOT register this module with `sys.modules` so as not to cause
|
||||||
|
confusion in the global environment. This way we have a valid proxy
|
||||||
|
module for our own use, but it lives _exclusively_ within lance.
|
||||||
|
|
||||||
|
"""
|
||||||
|
|
||||||
|
__lazy__ = True
|
||||||
|
|
||||||
|
_mod_pfx: ClassVar[dict[str, str]] = {
|
||||||
|
"numpy": "np.",
|
||||||
|
"pandas": "pd.",
|
||||||
|
"polars": "pl.",
|
||||||
|
"torch": "torch.",
|
||||||
|
"tensorflow": "tf.",
|
||||||
|
"ray": "ray.",
|
||||||
|
}
|
||||||
|
|
||||||
|
def __init__(
|
||||||
|
self,
|
||||||
|
module_name: str,
|
||||||
|
*,
|
||||||
|
module_available: bool,
|
||||||
|
) -> None:
|
||||||
|
"""
|
||||||
|
Initialise lazy-loading proxy module.
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
module_name : str
|
||||||
|
the name of the module to lazy-load (if available).
|
||||||
|
|
||||||
|
module_available : bool
|
||||||
|
indicate if the referenced module is actually available (we will proxy it
|
||||||
|
in both cases, but raise a helpful error when invoked if it doesn't exist).
|
||||||
|
|
||||||
|
"""
|
||||||
|
self._module_available = module_available
|
||||||
|
self._module_name = module_name
|
||||||
|
self._globals = globals()
|
||||||
|
super().__init__(module_name)
|
||||||
|
|
||||||
|
def _import(self) -> ModuleType:
|
||||||
|
# import the referenced module, replacing the proxy in this module's globals
|
||||||
|
module = import_module(self.__name__)
|
||||||
|
self._globals[self._module_name] = module
|
||||||
|
self.__dict__.update(module.__dict__)
|
||||||
|
return module
|
||||||
|
|
||||||
|
def __getattr__(self, attr: Any) -> Any:
|
||||||
|
# have "hasattr('__wrapped__')" return False without triggering import
|
||||||
|
# (it's for decorators, not modules, but keeps "make doctest" happy)
|
||||||
|
if attr == "__wrapped__":
|
||||||
|
raise AttributeError(
|
||||||
|
f"{self._module_name!r} object has no attribute {attr!r}"
|
||||||
|
)
|
||||||
|
|
||||||
|
# accessing the proxy module's attributes triggers import of the real thing
|
||||||
|
if self._module_available:
|
||||||
|
# import the module and return the requested attribute
|
||||||
|
module = self._import()
|
||||||
|
return getattr(module, attr)
|
||||||
|
|
||||||
|
# user has not installed the proxied/lazy module
|
||||||
|
elif attr == "__name__":
|
||||||
|
return self._module_name
|
||||||
|
elif re.match(r"^__\w+__$", attr) and attr != "__version__":
|
||||||
|
# allow some minimal introspection on private module
|
||||||
|
# attrs to avoid unnecessary error-handling elsewhere
|
||||||
|
return None
|
||||||
|
else:
|
||||||
|
# all other attribute access raises a helpful exception
|
||||||
|
pfx = self._mod_pfx.get(self._module_name, "")
|
||||||
|
raise ModuleNotFoundError(
|
||||||
|
f"{pfx}{attr} requires {self._module_name!r} module to be installed"
|
||||||
|
) from None
|
||||||
|
|
||||||
|
|
||||||
|
def _lazy_import(module_name: str) -> tuple[ModuleType, bool]:
|
||||||
|
"""
|
||||||
|
Lazy import the given module; avoids up-front import costs.
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
module_name : str
|
||||||
|
name of the module to import, eg: "polars".
|
||||||
|
|
||||||
|
Notes
|
||||||
|
-----
|
||||||
|
If the requested module is not available (eg: has not been installed), a proxy
|
||||||
|
module is created in its place, which raises an exception on any attribute
|
||||||
|
access. This allows for import and use as normal, without requiring explicit
|
||||||
|
guard conditions - if the module is never used, no exception occurs; if it
|
||||||
|
is, then a helpful exception is raised.
|
||||||
|
|
||||||
|
Returns
|
||||||
|
-------
|
||||||
|
tuple of (Module, bool)
|
||||||
|
A lazy-loading module and a boolean indicating if the requested/underlying
|
||||||
|
module exists (if not, the returned module is a proxy).
|
||||||
|
|
||||||
|
"""
|
||||||
|
# check if module is LOADED
|
||||||
|
if module_name in sys.modules:
|
||||||
|
return sys.modules[module_name], True
|
||||||
|
|
||||||
|
# check if module is AVAILABLE
|
||||||
|
try:
|
||||||
|
module_spec = find_spec(module_name)
|
||||||
|
module_available = not (module_spec is None or module_spec.loader is None)
|
||||||
|
except ModuleNotFoundError:
|
||||||
|
module_available = False
|
||||||
|
|
||||||
|
# create lazy/proxy module that imports the real one on first use
|
||||||
|
# (or raises an explanatory ModuleNotFoundError if not available)
|
||||||
|
return (
|
||||||
|
_LazyModule(
|
||||||
|
module_name=module_name,
|
||||||
|
module_available=module_available,
|
||||||
|
),
|
||||||
|
module_available,
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
if TYPE_CHECKING:
|
||||||
|
import datasets
|
||||||
|
import numpy
|
||||||
|
import pandas
|
||||||
|
import polars
|
||||||
|
import ray
|
||||||
|
import tensorflow
|
||||||
|
import torch
|
||||||
|
else:
|
||||||
|
# heavy/optional third party libs
|
||||||
|
numpy, _NUMPY_AVAILABLE = _lazy_import("numpy")
|
||||||
|
pandas, _PANDAS_AVAILABLE = _lazy_import("pandas")
|
||||||
|
polars, _POLARS_AVAILABLE = _lazy_import("polars")
|
||||||
|
torch, _TORCH_AVAILABLE = _lazy_import("torch")
|
||||||
|
datasets, _HUGGING_FACE_AVAILABLE = _lazy_import("datasets")
|
||||||
|
tensorflow, _TENSORFLOW_AVAILABLE = _lazy_import("tensorflow")
|
||||||
|
ray, _RAY_AVAILABLE = _lazy_import("ray")
|
||||||
|
|
||||||
|
|
||||||
|
@lru_cache(maxsize=None)
|
||||||
|
def _might_be(cls: type, type_: str) -> bool:
|
||||||
|
# infer whether the given class "might" be associated with the given
|
||||||
|
# module (in which case it's reasonable to do a real isinstance check)
|
||||||
|
try:
|
||||||
|
return any(f"{type_}." in str(o) for o in cls.mro())
|
||||||
|
except TypeError:
|
||||||
|
return False
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_numpy(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _NUMPY_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "numpy"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_pandas(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _PANDAS_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "pandas"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_polars(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _POLARS_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "polars"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_torch(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _TORCH_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "torch"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_hugging_face(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _HUGGING_FACE_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "datasets"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_tensorflow(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _TENSORFLOW_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "tensorflow"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _check_for_ray(obj: Any, *, check_type: bool = True) -> bool:
|
||||||
|
return _RAY_AVAILABLE and _might_be(
|
||||||
|
cast(Hashable, type(obj) if check_type else obj), "ray"
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
__all__ = [
|
||||||
|
# lazy-load third party libs
|
||||||
|
"datasets",
|
||||||
|
"numpy",
|
||||||
|
"pandas",
|
||||||
|
"polars",
|
||||||
|
"ray",
|
||||||
|
"tensorflow",
|
||||||
|
"torch",
|
||||||
|
# lazy utilities
|
||||||
|
"_check_for_hugging_face",
|
||||||
|
"_check_for_numpy",
|
||||||
|
"_check_for_pandas",
|
||||||
|
"_check_for_polars",
|
||||||
|
"_check_for_tensorflow",
|
||||||
|
"_check_for_torch",
|
||||||
|
"_check_for_ray",
|
||||||
|
"_LazyModule",
|
||||||
|
# exported flags/guards
|
||||||
|
"_NUMPY_AVAILABLE",
|
||||||
|
"_PANDAS_AVAILABLE",
|
||||||
|
"_POLARS_AVAILABLE",
|
||||||
|
"_TORCH_AVAILABLE",
|
||||||
|
"_HUGGING_FACE_AVAILABLE",
|
||||||
|
"_TENSORFLOW_AVAILABLE",
|
||||||
|
"_RAY_AVAILABLE",
|
||||||
|
]
|
||||||
@@ -83,7 +83,108 @@ class FTS:
|
|||||||
|
|
||||||
|
|
||||||
class HnswPq:
|
class HnswPq:
|
||||||
"""Describe a Hnswpq index configuration."""
|
"""Describe a HNSW-PQ index configuration.
|
||||||
|
|
||||||
|
HNSW-PQ stands for Hierarchical Navigable Small World - Product Quantization.
|
||||||
|
It is a variant of the HNSW algorithm that uses product quantization to compress
|
||||||
|
the vectors. To create an HNSW-PQ index, you can specify the following parameters:
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
|
||||||
|
distance_type: str, default "L2"
|
||||||
|
|
||||||
|
The distance metric used to train the index.
|
||||||
|
|
||||||
|
The following distance types are available:
|
||||||
|
|
||||||
|
"l2" - Euclidean distance. This is a very common distance metric that
|
||||||
|
accounts for both magnitude and direction when determining the distance
|
||||||
|
between vectors. L2 distance has a range of [0, ∞).
|
||||||
|
|
||||||
|
"cosine" - Cosine distance. Cosine distance is a distance metric
|
||||||
|
calculated from the cosine similarity between two vectors. Cosine
|
||||||
|
similarity is a measure of similarity between two non-zero vectors of an
|
||||||
|
inner product space. It is defined to equal the cosine of the angle
|
||||||
|
between them. Unlike L2, the cosine distance is not affected by the
|
||||||
|
magnitude of the vectors. Cosine distance has a range of [0, 2].
|
||||||
|
|
||||||
|
"dot" - Dot product. Dot distance is the dot product of two vectors. Dot
|
||||||
|
distance has a range of (-∞, ∞). If the vectors are normalized (i.e. their
|
||||||
|
L2 norm is 1), then dot distance is equivalent to the cosine distance.
|
||||||
|
|
||||||
|
num_partitions, default sqrt(num_rows)
|
||||||
|
|
||||||
|
The number of IVF partitions to create.
|
||||||
|
|
||||||
|
For HNSW, we recommend a small number of partitions. Setting this to 1 works
|
||||||
|
well for most tables. For very large tables, training just one HNSW graph
|
||||||
|
will require too much memory. Each partition becomes its own HNSW graph, so
|
||||||
|
setting this value higher reduces the peak memory use of training.
|
||||||
|
|
||||||
|
num_sub_vectors, default is vector dimension / 16
|
||||||
|
|
||||||
|
Number of sub-vectors of PQ.
|
||||||
|
|
||||||
|
This value controls how much the vector is compressed during the
|
||||||
|
quantization step. The more sub vectors there are the less the vector is
|
||||||
|
compressed. The default is the dimension of the vector divided by 16.
|
||||||
|
If the dimension is not evenly divisible by 16 we use the dimension
|
||||||
|
divided by 8.
|
||||||
|
|
||||||
|
The above two cases are highly preferred. Having 8 or 16 values per
|
||||||
|
subvector allows us to use efficient SIMD instructions.
|
||||||
|
|
||||||
|
If the dimension is not visible by 8 then we use 1 subvector. This is not
|
||||||
|
ideal and will likely result in poor performance.
|
||||||
|
|
||||||
|
max_iterations, default 50
|
||||||
|
|
||||||
|
Max iterations to train kmeans.
|
||||||
|
|
||||||
|
When training an IVF index we use kmeans to calculate the partitions. This
|
||||||
|
parameter controls how many iterations of kmeans to run.
|
||||||
|
|
||||||
|
Increasing this might improve the quality of the index but in most cases the
|
||||||
|
parameter is unused because kmeans will converge with fewer iterations. The
|
||||||
|
parameter is only used in cases where kmeans does not appear to converge. In
|
||||||
|
those cases it is unlikely that setting this larger will lead to the index
|
||||||
|
converging anyways.
|
||||||
|
|
||||||
|
sample_rate, default 256
|
||||||
|
|
||||||
|
The rate used to calculate the number of training vectors for kmeans.
|
||||||
|
|
||||||
|
When an IVF index is trained, we need to calculate partitions. These are
|
||||||
|
groups of vectors that are similar to each other. To do this we use an
|
||||||
|
algorithm called kmeans.
|
||||||
|
|
||||||
|
Running kmeans on a large dataset can be slow. To speed this up we
|
||||||
|
run kmeans on a random sample of the data. This parameter controls the
|
||||||
|
size of the sample. The total number of vectors used to train the index
|
||||||
|
is `sample_rate * num_partitions`.
|
||||||
|
|
||||||
|
Increasing this value might improve the quality of the index but in
|
||||||
|
most cases the default should be sufficient.
|
||||||
|
|
||||||
|
m, default 20
|
||||||
|
|
||||||
|
The number of neighbors to select for each vector in the HNSW graph.
|
||||||
|
|
||||||
|
This value controls the tradeoff between search speed and accuracy.
|
||||||
|
The higher the value the more accurate the search but the slower it will be.
|
||||||
|
|
||||||
|
ef_construction, default 300
|
||||||
|
|
||||||
|
The number of candidates to evaluate during the construction of the HNSW graph.
|
||||||
|
|
||||||
|
This value controls the tradeoff between build speed and accuracy.
|
||||||
|
The higher the value the more accurate the build but the slower it will be.
|
||||||
|
150 to 300 is the typical range. 100 is a minimum for good quality search
|
||||||
|
results. In most cases, there is no benefit to setting this higher than 500.
|
||||||
|
This value should be set to a value that is not less than `ef` in the
|
||||||
|
search phase.
|
||||||
|
"""
|
||||||
|
|
||||||
def __init__(
|
def __init__(
|
||||||
self,
|
self,
|
||||||
@@ -108,7 +209,93 @@ class HnswPq:
|
|||||||
|
|
||||||
|
|
||||||
class HnswSq:
|
class HnswSq:
|
||||||
"""Describe a HNSW-SQ index configuration."""
|
"""Describe a HNSW-SQ index configuration.
|
||||||
|
|
||||||
|
HNSW-SQ stands for Hierarchical Navigable Small World - Scalar Quantization.
|
||||||
|
It is a variant of the HNSW algorithm that uses scalar quantization to compress
|
||||||
|
the vectors.
|
||||||
|
|
||||||
|
Parameters
|
||||||
|
----------
|
||||||
|
|
||||||
|
distance_type: str, default "L2"
|
||||||
|
|
||||||
|
The distance metric used to train the index.
|
||||||
|
|
||||||
|
The following distance types are available:
|
||||||
|
|
||||||
|
"l2" - Euclidean distance. This is a very common distance metric that
|
||||||
|
accounts for both magnitude and direction when determining the distance
|
||||||
|
between vectors. L2 distance has a range of [0, ∞).
|
||||||
|
|
||||||
|
"cosine" - Cosine distance. Cosine distance is a distance metric
|
||||||
|
calculated from the cosine similarity between two vectors. Cosine
|
||||||
|
similarity is a measure of similarity between two non-zero vectors of an
|
||||||
|
inner product space. It is defined to equal the cosine of the angle
|
||||||
|
between them. Unlike L2, the cosine distance is not affected by the
|
||||||
|
magnitude of the vectors. Cosine distance has a range of [0, 2].
|
||||||
|
|
||||||
|
"dot" - Dot product. Dot distance is the dot product of two vectors. Dot
|
||||||
|
distance has a range of (-∞, ∞). If the vectors are normalized (i.e. their
|
||||||
|
L2 norm is 1), then dot distance is equivalent to the cosine distance.
|
||||||
|
|
||||||
|
num_partitions, default sqrt(num_rows)
|
||||||
|
|
||||||
|
The number of IVF partitions to create.
|
||||||
|
|
||||||
|
For HNSW, we recommend a small number of partitions. Setting this to 1 works
|
||||||
|
well for most tables. For very large tables, training just one HNSW graph
|
||||||
|
will require too much memory. Each partition becomes its own HNSW graph, so
|
||||||
|
setting this value higher reduces the peak memory use of training.
|
||||||
|
|
||||||
|
max_iterations, default 50
|
||||||
|
|
||||||
|
Max iterations to train kmeans.
|
||||||
|
|
||||||
|
When training an IVF index we use kmeans to calculate the partitions.
|
||||||
|
This parameter controls how many iterations of kmeans to run.
|
||||||
|
|
||||||
|
Increasing this might improve the quality of the index but in most cases
|
||||||
|
the parameter is unused because kmeans will converge with fewer iterations.
|
||||||
|
The parameter is only used in cases where kmeans does not appear to converge.
|
||||||
|
In those cases it is unlikely that setting this larger will lead to
|
||||||
|
the index converging anyways.
|
||||||
|
|
||||||
|
sample_rate, default 256
|
||||||
|
|
||||||
|
The rate used to calculate the number of training vectors for kmeans.
|
||||||
|
|
||||||
|
When an IVF index is trained, we need to calculate partitions. These
|
||||||
|
are groups of vectors that are similar to each other. To do this
|
||||||
|
we use an algorithm called kmeans.
|
||||||
|
|
||||||
|
Running kmeans on a large dataset can be slow. To speed this up we
|
||||||
|
run kmeans on a random sample of the data. This parameter controls the
|
||||||
|
size of the sample. The total number of vectors used to train the index
|
||||||
|
is `sample_rate * num_partitions`.
|
||||||
|
|
||||||
|
Increasing this value might improve the quality of the index but in
|
||||||
|
most cases the default should be sufficient.
|
||||||
|
|
||||||
|
m, default 20
|
||||||
|
|
||||||
|
The number of neighbors to select for each vector in the HNSW graph.
|
||||||
|
|
||||||
|
This value controls the tradeoff between search speed and accuracy.
|
||||||
|
The higher the value the more accurate the search but the slower it will be.
|
||||||
|
|
||||||
|
ef_construction, default 300
|
||||||
|
|
||||||
|
The number of candidates to evaluate during the construction of the HNSW graph.
|
||||||
|
|
||||||
|
This value controls the tradeoff between build speed and accuracy.
|
||||||
|
The higher the value the more accurate the build but the slower it will be.
|
||||||
|
150 to 300 is the typical range. 100 is a minimum for good quality search
|
||||||
|
results. In most cases, there is no benefit to setting this higher than 500.
|
||||||
|
This value should be set to a value that is not less than `ef` in the search
|
||||||
|
phase.
|
||||||
|
|
||||||
|
"""
|
||||||
|
|
||||||
def __init__(
|
def __init__(
|
||||||
self,
|
self,
|
||||||
|
|||||||
@@ -79,6 +79,13 @@ class RestfulLanceDBClient:
|
|||||||
or f"https://{self.db_name}.{self.region}.api.lancedb.com"
|
or f"https://{self.db_name}.{self.region}.api.lancedb.com"
|
||||||
)
|
)
|
||||||
|
|
||||||
|
def __enter__(self):
|
||||||
|
return self
|
||||||
|
|
||||||
|
def __exit__(self, exc_type, exc_value, traceback):
|
||||||
|
self.close()
|
||||||
|
return False # Do not suppress exceptions
|
||||||
|
|
||||||
def close(self):
|
def close(self):
|
||||||
self.session.close()
|
self.session.close()
|
||||||
self.closed = True
|
self.closed = True
|
||||||
|
|||||||
@@ -32,6 +32,9 @@ class AnswerdotaiRerankers(Reranker):
|
|||||||
The name of the column to use as input to the cross encoder model.
|
The name of the column to use as input to the cross encoder model.
|
||||||
return_score : str, default "relevance"
|
return_score : str, default "relevance"
|
||||||
options are "relevance" or "all". Only "relevance" is supported for now.
|
options are "relevance" or "all". Only "relevance" is supported for now.
|
||||||
|
**kwargs
|
||||||
|
Additional keyword arguments to pass to the model. For example, 'device'.
|
||||||
|
See AnswerDotAI/rerankers for more information.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(
|
def __init__(
|
||||||
@@ -40,13 +43,14 @@ class AnswerdotaiRerankers(Reranker):
|
|||||||
model_name: str = "answerdotai/answerai-colbert-small-v1",
|
model_name: str = "answerdotai/answerai-colbert-small-v1",
|
||||||
column: str = "text",
|
column: str = "text",
|
||||||
return_score="relevance",
|
return_score="relevance",
|
||||||
|
**kwargs,
|
||||||
):
|
):
|
||||||
super().__init__(return_score)
|
super().__init__(return_score)
|
||||||
self.column = column
|
self.column = column
|
||||||
rerankers = attempt_import_or_raise(
|
rerankers = attempt_import_or_raise(
|
||||||
"rerankers"
|
"rerankers"
|
||||||
) # import here for faster ops later
|
) # import here for faster ops later
|
||||||
self.reranker = rerankers.Reranker(model_name, model_type)
|
self.reranker = rerankers.Reranker(model_name, model_type, **kwargs)
|
||||||
|
|
||||||
def _rerank(self, result_set: pa.Table, query: str):
|
def _rerank(self, result_set: pa.Table, query: str):
|
||||||
docs = result_set[self.column].to_pylist()
|
docs = result_set[self.column].to_pylist()
|
||||||
|
|||||||
@@ -26,6 +26,9 @@ class ColbertReranker(AnswerdotaiRerankers):
|
|||||||
The name of the column to use as input to the cross encoder model.
|
The name of the column to use as input to the cross encoder model.
|
||||||
return_score : str, default "relevance"
|
return_score : str, default "relevance"
|
||||||
options are "relevance" or "all". Only "relevance" is supported for now.
|
options are "relevance" or "all". Only "relevance" is supported for now.
|
||||||
|
**kwargs
|
||||||
|
Additional keyword arguments to pass to the model, for example, 'device'.
|
||||||
|
See AnswerDotAI/rerankers for more information.
|
||||||
"""
|
"""
|
||||||
|
|
||||||
def __init__(
|
def __init__(
|
||||||
@@ -33,10 +36,12 @@ class ColbertReranker(AnswerdotaiRerankers):
|
|||||||
model_name: str = "colbert-ir/colbertv2.0",
|
model_name: str = "colbert-ir/colbertv2.0",
|
||||||
column: str = "text",
|
column: str = "text",
|
||||||
return_score="relevance",
|
return_score="relevance",
|
||||||
|
**kwargs,
|
||||||
):
|
):
|
||||||
super().__init__(
|
super().__init__(
|
||||||
model_type="colbert",
|
model_type="colbert",
|
||||||
model_name=model_name,
|
model_name=model_name,
|
||||||
column=column,
|
column=column,
|
||||||
return_score=return_score,
|
return_score=return_score,
|
||||||
|
**kwargs,
|
||||||
)
|
)
|
||||||
|
|||||||
@@ -19,10 +19,12 @@ from typing import (
|
|||||||
Optional,
|
Optional,
|
||||||
Tuple,
|
Tuple,
|
||||||
Union,
|
Union,
|
||||||
|
overload,
|
||||||
)
|
)
|
||||||
from urllib.parse import urlparse
|
from urllib.parse import urlparse
|
||||||
|
|
||||||
import lance
|
import lance
|
||||||
|
from .dependencies import _check_for_pandas
|
||||||
import numpy as np
|
import numpy as np
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
import pyarrow.compute as pc
|
import pyarrow.compute as pc
|
||||||
@@ -35,7 +37,16 @@ from .common import DATA, VEC, VECTOR_COLUMN_NAME
|
|||||||
from .embeddings import EmbeddingFunctionConfig, EmbeddingFunctionRegistry
|
from .embeddings import EmbeddingFunctionConfig, EmbeddingFunctionRegistry
|
||||||
from .merge import LanceMergeInsertBuilder
|
from .merge import LanceMergeInsertBuilder
|
||||||
from .pydantic import LanceModel, model_to_dict
|
from .pydantic import LanceModel, model_to_dict
|
||||||
from .query import AsyncQuery, AsyncVectorQuery, LanceQueryBuilder, Query
|
from .query import (
|
||||||
|
AsyncQuery,
|
||||||
|
AsyncVectorQuery,
|
||||||
|
LanceEmptyQueryBuilder,
|
||||||
|
LanceFtsQueryBuilder,
|
||||||
|
LanceHybridQueryBuilder,
|
||||||
|
LanceQueryBuilder,
|
||||||
|
LanceVectorQueryBuilder,
|
||||||
|
Query,
|
||||||
|
)
|
||||||
from .util import (
|
from .util import (
|
||||||
fs_from_uri,
|
fs_from_uri,
|
||||||
get_uri_scheme,
|
get_uri_scheme,
|
||||||
@@ -53,38 +64,25 @@ if TYPE_CHECKING:
|
|||||||
from .db import LanceDBConnection
|
from .db import LanceDBConnection
|
||||||
from .index import BTree, IndexConfig, IvfPq, Bitmap, LabelList, FTS
|
from .index import BTree, IndexConfig, IvfPq, Bitmap, LabelList, FTS
|
||||||
|
|
||||||
|
|
||||||
pd = safe_import_pandas()
|
pd = safe_import_pandas()
|
||||||
pl = safe_import_polars()
|
pl = safe_import_polars()
|
||||||
|
|
||||||
|
QueryType = Literal["vector", "fts", "hybrid", "auto"]
|
||||||
|
|
||||||
def _sanitize_data(
|
|
||||||
data,
|
def _coerce_to_table(data, schema: Optional[pa.Schema] = None) -> pa.Table:
|
||||||
schema: Optional[pa.Schema],
|
|
||||||
metadata: Optional[dict],
|
|
||||||
on_bad_vectors: str,
|
|
||||||
fill_value: Any,
|
|
||||||
):
|
|
||||||
if _check_for_hugging_face(data):
|
if _check_for_hugging_face(data):
|
||||||
# Huggingface datasets
|
# Huggingface datasets
|
||||||
from lance.dependencies import datasets
|
from lance.dependencies import datasets
|
||||||
|
|
||||||
if isinstance(data, datasets.dataset_dict.DatasetDict):
|
if isinstance(data, datasets.Dataset):
|
||||||
if schema is None:
|
|
||||||
schema = _schema_from_hf(data, schema)
|
|
||||||
data = _to_record_batch_generator(
|
|
||||||
_to_batches_with_split(data),
|
|
||||||
schema,
|
|
||||||
metadata,
|
|
||||||
on_bad_vectors,
|
|
||||||
fill_value,
|
|
||||||
)
|
|
||||||
elif isinstance(data, datasets.Dataset):
|
|
||||||
if schema is None:
|
if schema is None:
|
||||||
schema = data.features.arrow_schema
|
schema = data.features.arrow_schema
|
||||||
data = _to_record_batch_generator(
|
return pa.Table.from_batches(data.data.to_batches(), schema=schema)
|
||||||
data.data.to_batches(), schema, metadata, on_bad_vectors, fill_value
|
elif isinstance(data, datasets.dataset_dict.DatasetDict):
|
||||||
)
|
if schema is None:
|
||||||
|
schema = _schema_from_hf(data, schema)
|
||||||
|
return pa.Table.from_batches(_to_batches_with_split(data), schema=schema)
|
||||||
|
|
||||||
if isinstance(data, LanceModel):
|
if isinstance(data, LanceModel):
|
||||||
raise ValueError("Cannot add a single LanceModel to a table. Use a list.")
|
raise ValueError("Cannot add a single LanceModel to a table. Use a list.")
|
||||||
@@ -95,40 +93,68 @@ def _sanitize_data(
|
|||||||
if schema is None:
|
if schema is None:
|
||||||
schema = data[0].__class__.to_arrow_schema()
|
schema = data[0].__class__.to_arrow_schema()
|
||||||
data = [model_to_dict(d) for d in data]
|
data = [model_to_dict(d) for d in data]
|
||||||
data = pa.Table.from_pylist(data, schema=schema)
|
return pa.Table.from_pylist(data, schema=schema)
|
||||||
|
elif isinstance(data[0], pa.RecordBatch):
|
||||||
|
return pa.Table.from_batches(data, schema=schema)
|
||||||
else:
|
else:
|
||||||
data = pa.Table.from_pylist(data)
|
return pa.Table.from_pylist(data)
|
||||||
elif isinstance(data, dict):
|
elif isinstance(data, dict):
|
||||||
data = vec_to_table(data)
|
return vec_to_table(data)
|
||||||
elif pd is not None and isinstance(data, pd.DataFrame):
|
elif _check_for_pandas(data) and isinstance(data, pd.DataFrame):
|
||||||
data = pa.Table.from_pandas(data, preserve_index=False)
|
# Do not add schema here, since schema may contains the vector column
|
||||||
|
table = pa.Table.from_pandas(data, preserve_index=False)
|
||||||
# Do not serialize Pandas metadata
|
# Do not serialize Pandas metadata
|
||||||
meta = data.schema.metadata if data.schema.metadata is not None else {}
|
meta = table.schema.metadata if table.schema.metadata is not None else {}
|
||||||
meta = {k: v for k, v in meta.items() if k != b"pandas"}
|
meta = {k: v for k, v in meta.items() if k != b"pandas"}
|
||||||
data = data.replace_schema_metadata(meta)
|
return table.replace_schema_metadata(meta)
|
||||||
elif pl is not None and isinstance(data, pl.DataFrame):
|
elif isinstance(data, pa.Table):
|
||||||
data = data.to_arrow()
|
return data
|
||||||
|
elif isinstance(data, pa.RecordBatch):
|
||||||
if isinstance(data, pa.Table):
|
return pa.Table.from_batches([data])
|
||||||
if metadata:
|
elif isinstance(data, LanceDataset):
|
||||||
data = _append_vector_col(data, metadata, schema)
|
return data.scanner().to_table()
|
||||||
metadata.update(data.schema.metadata or {})
|
elif isinstance(data, pa.dataset.Dataset):
|
||||||
data = data.replace_schema_metadata(metadata)
|
return data.to_table()
|
||||||
data = _sanitize_schema(
|
elif isinstance(data, pa.dataset.Scanner):
|
||||||
data, schema=schema, on_bad_vectors=on_bad_vectors, fill_value=fill_value
|
return data.to_table()
|
||||||
)
|
elif isinstance(data, pa.RecordBatchReader):
|
||||||
if schema is None:
|
return data.read_all()
|
||||||
schema = data.schema
|
elif (
|
||||||
|
type(data).__module__.startswith("polars")
|
||||||
|
and data.__class__.__name__ == "DataFrame"
|
||||||
|
):
|
||||||
|
return data.to_arrow()
|
||||||
elif isinstance(data, Iterable):
|
elif isinstance(data, Iterable):
|
||||||
data = _to_record_batch_generator(
|
return _process_iterator(data, schema)
|
||||||
data, schema, metadata, on_bad_vectors, fill_value
|
|
||||||
)
|
|
||||||
if schema is None:
|
|
||||||
data, schema = _generator_to_data_and_schema(data)
|
|
||||||
if schema is None:
|
|
||||||
raise ValueError("Cannot infer schema from generator data")
|
|
||||||
else:
|
else:
|
||||||
raise TypeError(f"Unsupported data type: {type(data)}")
|
raise TypeError(
|
||||||
|
f"Unknown data type {type(data)}. "
|
||||||
|
"Please check "
|
||||||
|
"https://lancedb.github.io/lancedb/python/python/ "
|
||||||
|
"to see supported types."
|
||||||
|
)
|
||||||
|
|
||||||
|
|
||||||
|
def _sanitize_data(
|
||||||
|
data: Any,
|
||||||
|
schema: Optional[pa.Schema] = None,
|
||||||
|
metadata: Optional[dict] = None, # embedding metadata
|
||||||
|
on_bad_vectors: str = "error",
|
||||||
|
fill_value: float = 0.0,
|
||||||
|
):
|
||||||
|
data = _coerce_to_table(data, schema)
|
||||||
|
|
||||||
|
if metadata:
|
||||||
|
data = _append_vector_col(data, metadata, schema)
|
||||||
|
metadata.update(data.schema.metadata or {})
|
||||||
|
data = data.replace_schema_metadata(metadata)
|
||||||
|
|
||||||
|
# TODO improve the logics in _sanitize_schema
|
||||||
|
data = _sanitize_schema(data, schema, on_bad_vectors, fill_value)
|
||||||
|
if schema is None:
|
||||||
|
schema = data.schema
|
||||||
|
|
||||||
|
_validate_schema(schema)
|
||||||
return data, schema
|
return data, schema
|
||||||
|
|
||||||
|
|
||||||
@@ -149,6 +175,9 @@ def sanitize_create_table(
|
|||||||
on_bad_vectors=on_bad_vectors,
|
on_bad_vectors=on_bad_vectors,
|
||||||
fill_value=fill_value,
|
fill_value=fill_value,
|
||||||
)
|
)
|
||||||
|
else:
|
||||||
|
if schema is not None:
|
||||||
|
data = pa.Table.from_pylist([], schema)
|
||||||
if schema is None:
|
if schema is None:
|
||||||
if data is None:
|
if data is None:
|
||||||
raise ValueError("Either data or schema must be provided")
|
raise ValueError("Either data or schema must be provided")
|
||||||
@@ -505,7 +534,7 @@ class Table(ABC):
|
|||||||
Only available with use_tantivy=False
|
Only available with use_tantivy=False
|
||||||
If False, do not store the positions of the terms in the text.
|
If False, do not store the positions of the terms in the text.
|
||||||
This can reduce the size of the index and improve indexing speed.
|
This can reduce the size of the index and improve indexing speed.
|
||||||
But it will not be possible to use phrase queries.
|
But it will raise an exception for phrase queries.
|
||||||
|
|
||||||
"""
|
"""
|
||||||
raise NotImplementedError
|
raise NotImplementedError
|
||||||
@@ -607,7 +636,7 @@ class Table(ABC):
|
|||||||
self,
|
self,
|
||||||
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
vector_column_name: Optional[str] = None,
|
vector_column_name: Optional[str] = None,
|
||||||
query_type: str = "auto",
|
query_type: QueryType = "auto",
|
||||||
ordering_field_name: Optional[str] = None,
|
ordering_field_name: Optional[str] = None,
|
||||||
fts_columns: Optional[Union[str, List[str]]] = None,
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
) -> LanceQueryBuilder:
|
) -> LanceQueryBuilder:
|
||||||
@@ -1487,11 +1516,51 @@ class LanceTable(Table):
|
|||||||
self.schema.metadata
|
self.schema.metadata
|
||||||
)
|
)
|
||||||
|
|
||||||
|
@overload
|
||||||
def search(
|
def search(
|
||||||
self,
|
self,
|
||||||
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
vector_column_name: Optional[str] = None,
|
vector_column_name: Optional[str] = None,
|
||||||
query_type: str = "auto",
|
query_type: Literal["vector"] = "vector",
|
||||||
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
|
) -> LanceVectorQueryBuilder: ...
|
||||||
|
|
||||||
|
@overload
|
||||||
|
def search(
|
||||||
|
self,
|
||||||
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
|
vector_column_name: Optional[str] = None,
|
||||||
|
query_type: Literal["fts"] = "fts",
|
||||||
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
|
) -> LanceFtsQueryBuilder: ...
|
||||||
|
|
||||||
|
@overload
|
||||||
|
def search(
|
||||||
|
self,
|
||||||
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
|
vector_column_name: Optional[str] = None,
|
||||||
|
query_type: Literal["hybrid"] = "hybrid",
|
||||||
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
|
) -> LanceHybridQueryBuilder: ...
|
||||||
|
|
||||||
|
@overload
|
||||||
|
def search(
|
||||||
|
self,
|
||||||
|
query: None = None,
|
||||||
|
vector_column_name: Optional[str] = None,
|
||||||
|
query_type: QueryType = "auto",
|
||||||
|
ordering_field_name: Optional[str] = None,
|
||||||
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
|
) -> LanceEmptyQueryBuilder: ...
|
||||||
|
|
||||||
|
def search(
|
||||||
|
self,
|
||||||
|
query: Optional[Union[VEC, str, "PIL.Image.Image", Tuple]] = None,
|
||||||
|
vector_column_name: Optional[str] = None,
|
||||||
|
query_type: QueryType = "auto",
|
||||||
ordering_field_name: Optional[str] = None,
|
ordering_field_name: Optional[str] = None,
|
||||||
fts_columns: Optional[Union[str, List[str]]] = None,
|
fts_columns: Optional[Union[str, List[str]]] = None,
|
||||||
) -> LanceQueryBuilder:
|
) -> LanceQueryBuilder:
|
||||||
@@ -2015,6 +2084,55 @@ def _sanitize_nans(data, fill_value, on_bad_vectors, vec_arr, vector_column_name
|
|||||||
return data
|
return data
|
||||||
|
|
||||||
|
|
||||||
|
def _validate_schema(schema: pa.Schema):
|
||||||
|
"""
|
||||||
|
Make sure the metadata is valid utf8
|
||||||
|
"""
|
||||||
|
if schema.metadata is not None:
|
||||||
|
_validate_metadata(schema.metadata)
|
||||||
|
|
||||||
|
|
||||||
|
def _validate_metadata(metadata: dict):
|
||||||
|
"""
|
||||||
|
Make sure the metadata values are valid utf8 (can be nested)
|
||||||
|
|
||||||
|
Raises ValueError if not valid utf8
|
||||||
|
"""
|
||||||
|
for k, v in metadata.items():
|
||||||
|
if isinstance(v, bytes):
|
||||||
|
try:
|
||||||
|
v.decode("utf8")
|
||||||
|
except UnicodeDecodeError:
|
||||||
|
raise ValueError(
|
||||||
|
f"Metadata key {k} is not valid utf8. "
|
||||||
|
"Consider base64 encode for generic binary metadata."
|
||||||
|
)
|
||||||
|
elif isinstance(v, dict):
|
||||||
|
_validate_metadata(v)
|
||||||
|
|
||||||
|
|
||||||
|
def _process_iterator(data: Iterable, schema: Optional[pa.Schema] = None) -> pa.Table:
|
||||||
|
batches = []
|
||||||
|
for batch in data:
|
||||||
|
batch_table = _coerce_to_table(batch, schema)
|
||||||
|
if schema is not None:
|
||||||
|
if batch_table.schema != schema:
|
||||||
|
try:
|
||||||
|
batch_table = batch_table.cast(schema)
|
||||||
|
except pa.lib.ArrowInvalid:
|
||||||
|
raise ValueError(
|
||||||
|
f"Input iterator yielded a batch with schema that "
|
||||||
|
f"does not match the expected schema.\nExpected:\n{schema}\n"
|
||||||
|
f"Got:\n{batch_table.schema}"
|
||||||
|
)
|
||||||
|
batches.append(batch_table)
|
||||||
|
|
||||||
|
if batches:
|
||||||
|
return pa.concat_tables(batches)
|
||||||
|
else:
|
||||||
|
raise ValueError("Input iterable is empty")
|
||||||
|
|
||||||
|
|
||||||
class AsyncTable:
|
class AsyncTable:
|
||||||
"""
|
"""
|
||||||
An AsyncTable is a collection of Records in a LanceDB Database.
|
An AsyncTable is a collection of Records in a LanceDB Database.
|
||||||
|
|||||||
@@ -219,6 +219,7 @@ def value_to_sql(value):
|
|||||||
|
|
||||||
@value_to_sql.register(str)
|
@value_to_sql.register(str)
|
||||||
def _(value: str):
|
def _(value: str):
|
||||||
|
value = value.replace("'", "''")
|
||||||
return f"'{value}'"
|
return f"'{value}'"
|
||||||
|
|
||||||
|
|
||||||
|
|||||||
@@ -594,7 +594,9 @@ async def test_create_in_v2_mode(tmp_path):
|
|||||||
db = await lancedb.connect_async(tmp_path)
|
db = await lancedb.connect_async(tmp_path)
|
||||||
|
|
||||||
# Create table in v1 mode
|
# Create table in v1 mode
|
||||||
tbl = await db.create_table("test", data=make_data(), schema=schema)
|
tbl = await db.create_table(
|
||||||
|
"test", data=make_data(), schema=schema, data_storage_version="legacy"
|
||||||
|
)
|
||||||
|
|
||||||
async def is_in_v2_mode(tbl):
|
async def is_in_v2_mode(tbl):
|
||||||
batches = await tbl.query().to_batches(max_batch_length=1024 * 10)
|
batches = await tbl.query().to_batches(max_batch_length=1024 * 10)
|
||||||
@@ -626,7 +628,9 @@ async def test_create_in_v2_mode(tmp_path):
|
|||||||
assert await is_in_v2_mode(tbl)
|
assert await is_in_v2_mode(tbl)
|
||||||
|
|
||||||
# Create empty table uses v1 mode by default
|
# Create empty table uses v1 mode by default
|
||||||
tbl = await db.create_table("test_empty_v2_default", data=None, schema=schema)
|
tbl = await db.create_table(
|
||||||
|
"test_empty_v2_default", data=None, schema=schema, data_storage_version="legacy"
|
||||||
|
)
|
||||||
await tbl.add(make_table())
|
await tbl.add(make_table())
|
||||||
|
|
||||||
assert not await is_in_v2_mode(tbl)
|
assert not await is_in_v2_mode(tbl)
|
||||||
|
|||||||
@@ -143,7 +143,7 @@ def test_create_index_with_stemming(tmp_path, table):
|
|||||||
@pytest.mark.parametrize("with_position", [True, False])
|
@pytest.mark.parametrize("with_position", [True, False])
|
||||||
def test_create_inverted_index(table, use_tantivy, with_position):
|
def test_create_inverted_index(table, use_tantivy, with_position):
|
||||||
if use_tantivy and not with_position:
|
if use_tantivy and not with_position:
|
||||||
pytest.skip("we don't support to build tantivy index without position")
|
pytest.skip("we don't support building a tantivy index without position")
|
||||||
table.create_fts_index("text", use_tantivy=use_tantivy, with_position=with_position)
|
table.create_fts_index("text", use_tantivy=use_tantivy, with_position=with_position)
|
||||||
|
|
||||||
|
|
||||||
|
|||||||
@@ -74,21 +74,23 @@ async def test_e2e_with_mock_server():
|
|||||||
await mock_server.start()
|
await mock_server.start()
|
||||||
|
|
||||||
try:
|
try:
|
||||||
client = RestfulLanceDBClient("lancedb+http://localhost:8111")
|
with RestfulLanceDBClient("lancedb+http://localhost:8111") as client:
|
||||||
df = (
|
df = (
|
||||||
await client.query(
|
await client.query(
|
||||||
"test_table",
|
"test_table",
|
||||||
VectorQuery(
|
VectorQuery(
|
||||||
vector=np.random.rand(128).tolist(),
|
vector=np.random.rand(128).tolist(),
|
||||||
k=10,
|
k=10,
|
||||||
_metric="L2",
|
_metric="L2",
|
||||||
columns=["id", "vector"],
|
columns=["id", "vector"],
|
||||||
),
|
),
|
||||||
)
|
)
|
||||||
).to_pandas()
|
).to_pandas()
|
||||||
|
|
||||||
assert "vector" in df.columns
|
assert "vector" in df.columns
|
||||||
assert "id" in df.columns
|
assert "id" in df.columns
|
||||||
|
|
||||||
|
assert client.closed
|
||||||
finally:
|
finally:
|
||||||
# make sure we don't leak resources
|
# make sure we don't leak resources
|
||||||
await mock_server.stop()
|
await mock_server.stop()
|
||||||
|
|||||||
@@ -1,15 +1,7 @@
|
|||||||
# Copyright 2023 LanceDB Developers
|
# SPDX-License-Identifier: Apache-2.0
|
||||||
#
|
# SPDX-FileCopyrightText: Copyright The LanceDB Authors
|
||||||
# Licensed under the Apache License, Version 2.0 (the "License");
|
|
||||||
# you may not use this file except in compliance with the License.
|
from unittest.mock import MagicMock
|
||||||
# You may obtain a copy of the License at
|
|
||||||
# http://www.apache.org/licenses/LICENSE-2.0
|
|
||||||
#
|
|
||||||
# Unless required by applicable law or agreed to in writing, software
|
|
||||||
# distributed under the License is distributed on an "AS IS" BASIS,
|
|
||||||
# WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
|
|
||||||
# See the License for the specific language governing permissions and
|
|
||||||
# limitations under the License.
|
|
||||||
|
|
||||||
import lancedb
|
import lancedb
|
||||||
import pyarrow as pa
|
import pyarrow as pa
|
||||||
@@ -39,3 +31,53 @@ def test_remote_db():
|
|||||||
table = conn["test"]
|
table = conn["test"]
|
||||||
table.schema = pa.schema([pa.field("vector", pa.list_(pa.float32(), 2))])
|
table.schema = pa.schema([pa.field("vector", pa.list_(pa.float32(), 2))])
|
||||||
table.search([1.0, 2.0]).to_pandas()
|
table.search([1.0, 2.0]).to_pandas()
|
||||||
|
|
||||||
|
|
||||||
|
def test_create_empty_table():
|
||||||
|
client = MagicMock()
|
||||||
|
conn = lancedb.connect("db://client-will-be-injected", api_key="fake")
|
||||||
|
|
||||||
|
conn._client = client
|
||||||
|
|
||||||
|
schema = pa.schema([pa.field("vector", pa.list_(pa.float32(), 2))])
|
||||||
|
|
||||||
|
client.post.return_value = {"status": "ok"}
|
||||||
|
table = conn.create_table("test", schema=schema)
|
||||||
|
assert table.name == "test"
|
||||||
|
assert client.post.call_args[0][0] == "/v1/table/test/create/"
|
||||||
|
|
||||||
|
json_schema = {
|
||||||
|
"fields": [
|
||||||
|
{
|
||||||
|
"name": "vector",
|
||||||
|
"nullable": True,
|
||||||
|
"type": {
|
||||||
|
"type": "fixed_size_list",
|
||||||
|
"fields": [
|
||||||
|
{"name": "item", "nullable": True, "type": {"type": "float"}}
|
||||||
|
],
|
||||||
|
"length": 2,
|
||||||
|
},
|
||||||
|
},
|
||||||
|
]
|
||||||
|
}
|
||||||
|
client.post.return_value = {"schema": json_schema}
|
||||||
|
assert table.schema == schema
|
||||||
|
assert client.post.call_args[0][0] == "/v1/table/test/describe/"
|
||||||
|
|
||||||
|
client.post.return_value = 0
|
||||||
|
assert table.count_rows(None) == 0
|
||||||
|
|
||||||
|
|
||||||
|
def test_create_table_with_recordbatches():
|
||||||
|
client = MagicMock()
|
||||||
|
conn = lancedb.connect("db://client-will-be-injected", api_key="fake")
|
||||||
|
|
||||||
|
conn._client = client
|
||||||
|
|
||||||
|
batch = pa.RecordBatch.from_arrays([pa.array([[1.0, 2.0], [3.0, 4.0]])], ["vector"])
|
||||||
|
|
||||||
|
client.post.return_value = {"status": "ok"}
|
||||||
|
table = conn.create_table("test", [batch], schema=batch.schema)
|
||||||
|
assert table.name == "test"
|
||||||
|
assert client.post.call_args[0][0] == "/v1/table/test/create/"
|
||||||
|
|||||||
@@ -64,6 +64,55 @@ def test_basic(db):
|
|||||||
assert table.to_lance().to_table() == ds.to_table()
|
assert table.to_lance().to_table() == ds.to_table()
|
||||||
|
|
||||||
|
|
||||||
|
def test_input_data_type(db, tmp_path):
|
||||||
|
schema = pa.schema(
|
||||||
|
[
|
||||||
|
pa.field("id", pa.int64()),
|
||||||
|
pa.field("name", pa.string()),
|
||||||
|
pa.field("age", pa.int32()),
|
||||||
|
]
|
||||||
|
)
|
||||||
|
|
||||||
|
data = {
|
||||||
|
"id": [1, 2, 3, 4, 5],
|
||||||
|
"name": ["Alice", "Bob", "Charlie", "David", "Eve"],
|
||||||
|
"age": [25, 30, 35, 40, 45],
|
||||||
|
}
|
||||||
|
record_batch = pa.RecordBatch.from_pydict(data, schema=schema)
|
||||||
|
pa_reader = pa.RecordBatchReader.from_batches(record_batch.schema, [record_batch])
|
||||||
|
pa_table = pa.Table.from_batches([record_batch])
|
||||||
|
|
||||||
|
def create_dataset(tmp_path):
|
||||||
|
path = os.path.join(tmp_path, "test_source_dataset")
|
||||||
|
pa.dataset.write_dataset(pa_table, path, format="parquet")
|
||||||
|
return pa.dataset.dataset(path, format="parquet")
|
||||||
|
|
||||||
|
pa_dataset = create_dataset(tmp_path)
|
||||||
|
pa_scanner = pa_dataset.scanner()
|
||||||
|
|
||||||
|
input_types = [
|
||||||
|
("RecordBatchReader", pa_reader),
|
||||||
|
("RecordBatch", record_batch),
|
||||||
|
("Table", pa_table),
|
||||||
|
("Dataset", pa_dataset),
|
||||||
|
("Scanner", pa_scanner),
|
||||||
|
]
|
||||||
|
for input_type, input_data in input_types:
|
||||||
|
table_name = f"test_{input_type.lower()}"
|
||||||
|
ds = LanceTable.create(db, table_name, data=input_data).to_lance()
|
||||||
|
assert ds.schema == schema
|
||||||
|
assert ds.count_rows() == 5
|
||||||
|
|
||||||
|
assert ds.schema.field("id").type == pa.int64()
|
||||||
|
assert ds.schema.field("name").type == pa.string()
|
||||||
|
assert ds.schema.field("age").type == pa.int32()
|
||||||
|
|
||||||
|
result_table = ds.to_table()
|
||||||
|
assert result_table.column("id").to_pylist() == data["id"]
|
||||||
|
assert result_table.column("name").to_pylist() == data["name"]
|
||||||
|
assert result_table.column("age").to_pylist() == data["age"]
|
||||||
|
|
||||||
|
|
||||||
@pytest.mark.asyncio
|
@pytest.mark.asyncio
|
||||||
async def test_close(db_async: AsyncConnection):
|
async def test_close(db_async: AsyncConnection):
|
||||||
table = await db_async.create_table("some_table", data=[{"id": 0}])
|
table = await db_async.create_table("some_table", data=[{"id": 0}])
|
||||||
@@ -274,7 +323,6 @@ def test_polars(db):
|
|||||||
|
|
||||||
|
|
||||||
def _add(table, schema):
|
def _add(table, schema):
|
||||||
# table = LanceTable(db, "test")
|
|
||||||
assert len(table) == 2
|
assert len(table) == 2
|
||||||
|
|
||||||
table.add([{"vector": [6.3, 100.5], "item": "new", "price": 30.0}])
|
table.add([{"vector": [6.3, 100.5], "item": "new", "price": 30.0}])
|
||||||
|
|||||||
@@ -15,7 +15,8 @@ import os
|
|||||||
import pathlib
|
import pathlib
|
||||||
|
|
||||||
import pytest
|
import pytest
|
||||||
from lancedb.util import get_uri_scheme, join_uri
|
import lancedb
|
||||||
|
from lancedb.util import get_uri_scheme, join_uri, value_to_sql
|
||||||
|
|
||||||
|
|
||||||
def test_normalize_uri():
|
def test_normalize_uri():
|
||||||
@@ -84,3 +85,29 @@ def test_local_join_uri_windows():
|
|||||||
assert joined == str(pathlib.Path(base) / "table.lance")
|
assert joined == str(pathlib.Path(base) / "table.lance")
|
||||||
joined = join_uri(pathlib.Path(base), "table.lance")
|
joined = join_uri(pathlib.Path(base), "table.lance")
|
||||||
assert joined == pathlib.Path(base) / "table.lance"
|
assert joined == pathlib.Path(base) / "table.lance"
|
||||||
|
|
||||||
|
|
||||||
|
def test_value_to_sql_string(tmp_path):
|
||||||
|
# Make sure we can convert Python string literals to SQL strings, even if
|
||||||
|
# they contain characters meaningful in SQL, such as ' and \.
|
||||||
|
values = ["anthony's", 'a "test" string', "anthony's \"favorite color\" wasn't red"]
|
||||||
|
expected_values = [
|
||||||
|
"'anthony''s'",
|
||||||
|
"'a \"test\" string'",
|
||||||
|
"'anthony''s \"favorite color\" wasn''t red'",
|
||||||
|
]
|
||||||
|
|
||||||
|
for value, expected in zip(values, expected_values):
|
||||||
|
assert value_to_sql(value) == expected
|
||||||
|
|
||||||
|
# Also test we can roundtrip those strings through update.
|
||||||
|
# This validates the query parser understands the strings we
|
||||||
|
# are creating.
|
||||||
|
db = lancedb.connect(tmp_path)
|
||||||
|
table = db.create_table(
|
||||||
|
"test",
|
||||||
|
[{"search": value, "replace": "something"} for value in values],
|
||||||
|
)
|
||||||
|
for value in values:
|
||||||
|
table.update(where=f"search = {value_to_sql(value)}", values={"replace": value})
|
||||||
|
assert table.to_pandas().query("search == @value")["replace"].item() == value
|
||||||
|
|||||||
@@ -1,11 +1,11 @@
|
|||||||
# Release process
|
# Release process
|
||||||
|
|
||||||
There are five total packages we release. Three are the `lancedb` packages
|
There are five total packages we release. Four are the `lancedb` packages
|
||||||
for Python, Rust, and Node.js. The other two are the legacy `vectordb`
|
for Python, Rust, Java, and Node.js. The other one is the legacy `vectordb`
|
||||||
packages for Rust and node.js.
|
package node.js.
|
||||||
|
|
||||||
The Python package is versioned and released separately from the Rust and Node.js
|
The Python package is versioned and released separately from the Rust, Java, and Node.js
|
||||||
ones. For Rust and Node.js, the release process is shared between `lancedb` and
|
ones. For Node.js the release process is shared between `lancedb` and
|
||||||
`vectordb` for now.
|
`vectordb` for now.
|
||||||
|
|
||||||
## Preview releases
|
## Preview releases
|
||||||
@@ -36,7 +36,10 @@ The release process uses a handful of GitHub actions to automate the process.
|
|||||||
│ └───────────┘ │
|
│ └───────────┘ │
|
||||||
│ └──►NPM Packages
|
│ └──►NPM Packages
|
||||||
│ ┌─────────────┐
|
│ ┌─────────────┐
|
||||||
└──────►│Cargo Publish├───►Cargo Release
|
├──────►│Cargo Publish├───►Cargo Release
|
||||||
|
│ └─────────────┘
|
||||||
|
│ ┌─────────────┐
|
||||||
|
└──────►│Maven Publish├───►Java Maven Repo Release
|
||||||
└─────────────┘
|
└─────────────┘
|
||||||
```
|
```
|
||||||
|
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb-node"
|
name = "lancedb-node"
|
||||||
version = "0.10.0-beta.1"
|
version = "0.10.0"
|
||||||
description = "Serverless, low-latency vector database for AI applications"
|
description = "Serverless, low-latency vector database for AI applications"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
|
|||||||
@@ -1,6 +1,6 @@
|
|||||||
[package]
|
[package]
|
||||||
name = "lancedb"
|
name = "lancedb"
|
||||||
version = "0.10.0-beta.1"
|
version = "0.10.0"
|
||||||
edition.workspace = true
|
edition.workspace = true
|
||||||
description = "LanceDB: A serverless, low-latency vector database for AI applications"
|
description = "LanceDB: A serverless, low-latency vector database for AI applications"
|
||||||
license.workspace = true
|
license.workspace = true
|
||||||
@@ -45,7 +45,8 @@ serde_json = { version = "1" }
|
|||||||
async-openai = { version = "0.20.0", optional = true }
|
async-openai = { version = "0.20.0", optional = true }
|
||||||
serde_with = { version = "3.8.1" }
|
serde_with = { version = "3.8.1" }
|
||||||
# For remote feature
|
# For remote feature
|
||||||
reqwest = { version = "0.11.24", features = ["gzip", "json"], optional = true }
|
reqwest = { version = "0.12.0", features = ["gzip", "json", "stream"], optional = true }
|
||||||
|
http = { version = "1", optional = true } # Matching what is in reqwest
|
||||||
polars-arrow = { version = ">=0.37,<0.40.0", optional = true }
|
polars-arrow = { version = ">=0.37,<0.40.0", optional = true }
|
||||||
polars = { version = ">=0.37,<0.40.0", optional = true }
|
polars = { version = ">=0.37,<0.40.0", optional = true }
|
||||||
hf-hub = { version = "0.3.2", optional = true }
|
hf-hub = { version = "0.3.2", optional = true }
|
||||||
@@ -65,10 +66,11 @@ aws-sdk-s3 = { version = "1.38.0" }
|
|||||||
aws-sdk-kms = { version = "1.37" }
|
aws-sdk-kms = { version = "1.37" }
|
||||||
aws-config = { version = "1.0" }
|
aws-config = { version = "1.0" }
|
||||||
aws-smithy-runtime = { version = "1.3" }
|
aws-smithy-runtime = { version = "1.3" }
|
||||||
|
http-body = "1" # Matching reqwest
|
||||||
|
|
||||||
[features]
|
[features]
|
||||||
default = []
|
default = []
|
||||||
remote = ["dep:reqwest"]
|
remote = ["dep:reqwest", "dep:http"]
|
||||||
fp16kernels = ["lance-linalg/fp16kernels"]
|
fp16kernels = ["lance-linalg/fp16kernels"]
|
||||||
s3-test = []
|
s3-test = []
|
||||||
openai = ["dep:async-openai", "dep:reqwest"]
|
openai = ["dep:async-openai", "dep:reqwest"]
|
||||||
|
|||||||
@@ -307,7 +307,7 @@ impl<const HAS_DATA: bool, T: IntoArrow> CreateTableBuilder<HAS_DATA, T> {
|
|||||||
|
|
||||||
/// Set the data storage version.
|
/// Set the data storage version.
|
||||||
///
|
///
|
||||||
/// The default is `LanceFileVersion::Legacy`.
|
/// The default is `LanceFileVersion::Stable`.
|
||||||
pub fn data_storage_version(mut self, data_storage_version: LanceFileVersion) -> Self {
|
pub fn data_storage_version(mut self, data_storage_version: LanceFileVersion) -> Self {
|
||||||
self.data_storage_version = Some(data_storage_version);
|
self.data_storage_version = Some(data_storage_version);
|
||||||
self
|
self
|
||||||
@@ -315,13 +315,9 @@ impl<const HAS_DATA: bool, T: IntoArrow> CreateTableBuilder<HAS_DATA, T> {
|
|||||||
|
|
||||||
/// Set to true to use the v1 format for data files
|
/// Set to true to use the v1 format for data files
|
||||||
///
|
///
|
||||||
/// This is currently defaulted to true and can be set to false to opt-in
|
/// This is set to false by default to enable the stable format.
|
||||||
/// to the new format. This should only be used for experimentation and
|
/// This should only be used for experimentation and
|
||||||
/// evaluation. The new format is still in beta and may change in ways that
|
/// evaluation. This option may be removed in the future releases.
|
||||||
/// are not backwards compatible.
|
|
||||||
///
|
|
||||||
/// Once the new format is stable, the default will change to `false` for
|
|
||||||
/// several releases and then eventually this option will be removed.
|
|
||||||
#[deprecated(since = "0.9.0", note = "use data_storage_version instead")]
|
#[deprecated(since = "0.9.0", note = "use data_storage_version instead")]
|
||||||
pub fn use_legacy_format(mut self, use_legacy_format: bool) -> Self {
|
pub fn use_legacy_format(mut self, use_legacy_format: bool) -> Self {
|
||||||
self.data_storage_version = if use_legacy_format {
|
self.data_storage_version = if use_legacy_format {
|
||||||
@@ -335,8 +331,8 @@ impl<const HAS_DATA: bool, T: IntoArrow> CreateTableBuilder<HAS_DATA, T> {
|
|||||||
|
|
||||||
#[derive(Clone, Debug)]
|
#[derive(Clone, Debug)]
|
||||||
pub struct OpenTableBuilder {
|
pub struct OpenTableBuilder {
|
||||||
parent: Arc<dyn ConnectionInternal>,
|
pub(crate) parent: Arc<dyn ConnectionInternal>,
|
||||||
name: String,
|
pub(crate) name: String,
|
||||||
index_cache_size: u32,
|
index_cache_size: u32,
|
||||||
lance_read_params: Option<ReadParams>,
|
lance_read_params: Option<ReadParams>,
|
||||||
}
|
}
|
||||||
@@ -1095,6 +1091,25 @@ impl ConnectionInternal for Database {
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[cfg(all(test, feature = "remote"))]
|
||||||
|
mod test_utils {
|
||||||
|
use super::*;
|
||||||
|
impl Connection {
|
||||||
|
pub fn new_with_handler<T>(
|
||||||
|
handler: impl Fn(reqwest::Request) -> http::Response<T> + Clone + Send + Sync + 'static,
|
||||||
|
) -> Self
|
||||||
|
where
|
||||||
|
T: Into<reqwest::Body>,
|
||||||
|
{
|
||||||
|
let internal = Arc::new(crate::remote::db::RemoteDatabase::new_mock(handler));
|
||||||
|
Self {
|
||||||
|
internal,
|
||||||
|
uri: "db://test".to_string(),
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
#[cfg(test)]
|
#[cfg(test)]
|
||||||
mod tests {
|
mod tests {
|
||||||
use arrow_schema::{DataType, Field, Schema};
|
use arrow_schema::{DataType, Field, Schema};
|
||||||
@@ -1208,9 +1223,9 @@ mod tests {
|
|||||||
assert_eq!(tables, vec!["table1".to_owned()]);
|
assert_eq!(tables, vec!["table1".to_owned()]);
|
||||||
}
|
}
|
||||||
|
|
||||||
fn make_data() -> impl RecordBatchReader + Send + 'static {
|
fn make_data() -> Box<dyn RecordBatchReader + Send + 'static> {
|
||||||
let id = Box::new(IncrementingInt32::new().named("id".to_string()));
|
let id = Box::new(IncrementingInt32::new().named("id".to_string()));
|
||||||
BatchGenerator::new().col(id).batches(10, 2000)
|
Box::new(BatchGenerator::new().col(id).batches(10, 2000))
|
||||||
}
|
}
|
||||||
|
|
||||||
#[tokio::test]
|
#[tokio::test]
|
||||||
@@ -1221,6 +1236,7 @@ mod tests {
|
|||||||
|
|
||||||
let tbl = db
|
let tbl = db
|
||||||
.create_table("v1_test", make_data())
|
.create_table("v1_test", make_data())
|
||||||
|
.data_storage_version(LanceFileVersion::Legacy)
|
||||||
.execute()
|
.execute()
|
||||||
.await
|
.await
|
||||||
.unwrap();
|
.unwrap();
|
||||||
|
|||||||
@@ -145,7 +145,7 @@ impl SentenceTransformersEmbeddingsBuilder {
|
|||||||
let device = self.device.unwrap_or(Device::Cpu);
|
let device = self.device.unwrap_or(Device::Cpu);
|
||||||
|
|
||||||
let repo = if let Some(revision) = self.revision {
|
let repo = if let Some(revision) = self.revision {
|
||||||
Repo::with_revision(model_id, RepoType::Model, revision.to_string())
|
Repo::with_revision(model_id, RepoType::Model, revision)
|
||||||
} else {
|
} else {
|
||||||
Repo::new(model_id, RepoType::Model)
|
Repo::new(model_id, RepoType::Model)
|
||||||
};
|
};
|
||||||
|
|||||||
@@ -59,9 +59,11 @@ pub enum Index {
|
|||||||
IvfPq(IvfPqIndexBuilder),
|
IvfPq(IvfPqIndexBuilder),
|
||||||
|
|
||||||
/// IVF-HNSW index with Product Quantization
|
/// IVF-HNSW index with Product Quantization
|
||||||
|
/// It is a variant of the HNSW algorithm that uses product quantization to compress the vectors.
|
||||||
IvfHnswPq(IvfHnswPqIndexBuilder),
|
IvfHnswPq(IvfHnswPqIndexBuilder),
|
||||||
|
|
||||||
/// IVF-HNSW index with Scalar Quantization
|
/// IVF-HNSW index with Scalar Quantization
|
||||||
|
/// It is a variant of the HNSW algorithm that uses scalar quantization to compress the vectors.
|
||||||
IvfHnswSq(IvfHnswSqIndexBuilder),
|
IvfHnswSq(IvfHnswSqIndexBuilder),
|
||||||
}
|
}
|
||||||
|
|
||||||
|
|||||||
@@ -21,3 +21,5 @@ pub mod client;
|
|||||||
pub mod db;
|
pub mod db;
|
||||||
pub mod table;
|
pub mod table;
|
||||||
pub mod util;
|
pub mod util;
|
||||||
|
|
||||||
|
const ARROW_STREAM_CONTENT_TYPE: &str = "application/vnd.apache.arrow.stream";
|
||||||
|
|||||||
@@ -12,7 +12,7 @@
|
|||||||
// See the License for the specific language governing permissions and
|
// See the License for the specific language governing permissions and
|
||||||
// limitations under the License.
|
// limitations under the License.
|
||||||
|
|
||||||
use std::time::Duration;
|
use std::{future::Future, time::Duration};
|
||||||
|
|
||||||
use reqwest::{
|
use reqwest::{
|
||||||
header::{HeaderMap, HeaderValue},
|
header::{HeaderMap, HeaderValue},
|
||||||
@@ -21,13 +21,66 @@ use reqwest::{
|
|||||||
|
|
||||||
use crate::error::{Error, Result};
|
use crate::error::{Error, Result};
|
||||||
|
|
||||||
|
// We use the `HttpSend` trait to abstract over the `reqwest::Client` so that
|
||||||
|
// we can mock responses in tests. Based on the patterns from this blog post:
|
||||||
|
// https://write.as/balrogboogie/testing-reqwest-based-clients
|
||||||
#[derive(Clone, Debug)]
|
#[derive(Clone, Debug)]
|
||||||
pub struct RestfulLanceDbClient {
|
pub struct RestfulLanceDbClient<S: HttpSend = Sender> {
|
||||||
client: reqwest::Client,
|
client: reqwest::Client,
|
||||||
host: String,
|
host: String,
|
||||||
|
sender: S,
|
||||||
}
|
}
|
||||||
|
|
||||||
impl RestfulLanceDbClient {
|
pub trait HttpSend: Clone + Send + Sync + std::fmt::Debug + 'static {
|
||||||
|
fn send(&self, req: RequestBuilder) -> impl Future<Output = Result<Response>> + Send;
|
||||||
|
}
|
||||||
|
|
||||||
|
// Default implementation of HttpSend which sends the request normally with reqwest
|
||||||
|
#[derive(Clone, Debug)]
|
||||||
|
pub struct Sender;
|
||||||
|
impl HttpSend for Sender {
|
||||||
|
async fn send(&self, request: reqwest::RequestBuilder) -> Result<reqwest::Response> {
|
||||||
|
Ok(request.send().await?)
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
impl RestfulLanceDbClient<Sender> {
|
||||||
|
pub fn try_new(
|
||||||
|
db_url: &str,
|
||||||
|
api_key: &str,
|
||||||
|
region: &str,
|
||||||
|
host_override: Option<String>,
|
||||||
|
) -> Result<Self> {
|
||||||
|
let parsed_url = url::Url::parse(db_url)?;
|
||||||
|
debug_assert_eq!(parsed_url.scheme(), "db");
|
||||||
|
if !parsed_url.has_host() {
|
||||||
|
return Err(Error::Http {
|
||||||
|
message: format!("Invalid database URL (missing host) '{}'", db_url),
|
||||||
|
});
|
||||||
|
}
|
||||||
|
let db_name = parsed_url.host_str().unwrap();
|
||||||
|
let client = reqwest::Client::builder()
|
||||||
|
.timeout(Duration::from_secs(30))
|
||||||
|
.default_headers(Self::default_headers(
|
||||||
|
api_key,
|
||||||
|
region,
|
||||||
|
db_name,
|
||||||
|
host_override.is_some(),
|
||||||
|
)?)
|
||||||
|
.build()?;
|
||||||
|
let host = match host_override {
|
||||||
|
Some(host_override) => host_override,
|
||||||
|
None => format!("https://{}.{}.api.lancedb.com", db_name, region),
|
||||||
|
};
|
||||||
|
Ok(Self {
|
||||||
|
client,
|
||||||
|
host,
|
||||||
|
sender: Sender,
|
||||||
|
})
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
impl<S: HttpSend> RestfulLanceDbClient<S> {
|
||||||
pub fn host(&self) -> &str {
|
pub fn host(&self) -> &str {
|
||||||
&self.host
|
&self.host
|
||||||
}
|
}
|
||||||
@@ -66,36 +119,6 @@ impl RestfulLanceDbClient {
|
|||||||
Ok(headers)
|
Ok(headers)
|
||||||
}
|
}
|
||||||
|
|
||||||
pub fn try_new(
|
|
||||||
db_url: &str,
|
|
||||||
api_key: &str,
|
|
||||||
region: &str,
|
|
||||||
host_override: Option<String>,
|
|
||||||
) -> Result<Self> {
|
|
||||||
let parsed_url = url::Url::parse(db_url)?;
|
|
||||||
debug_assert_eq!(parsed_url.scheme(), "db");
|
|
||||||
if !parsed_url.has_host() {
|
|
||||||
return Err(Error::Http {
|
|
||||||
message: format!("Invalid database URL (missing host) '{}'", db_url),
|
|
||||||
});
|
|
||||||
}
|
|
||||||
let db_name = parsed_url.host_str().unwrap();
|
|
||||||
let client = reqwest::Client::builder()
|
|
||||||
.timeout(Duration::from_secs(30))
|
|
||||||
.default_headers(Self::default_headers(
|
|
||||||
api_key,
|
|
||||||
region,
|
|
||||||
db_name,
|
|
||||||
host_override.is_some(),
|
|
||||||
)?)
|
|
||||||
.build()?;
|
|
||||||
let host = match host_override {
|
|
||||||
Some(host_override) => host_override,
|
|
||||||
None => format!("https://{}.{}.api.lancedb.com", db_name, region),
|
|
||||||
};
|
|
||||||
Ok(Self { client, host })
|
|
||||||
}
|
|
||||||
|
|
||||||
pub fn get(&self, uri: &str) -> RequestBuilder {
|
pub fn get(&self, uri: &str) -> RequestBuilder {
|
||||||
let full_uri = format!("{}{}", self.host, uri);
|
let full_uri = format!("{}{}", self.host, uri);
|
||||||
self.client.get(full_uri)
|
self.client.get(full_uri)
|
||||||
@@ -106,6 +129,10 @@ impl RestfulLanceDbClient {
|
|||||||
self.client.post(full_uri)
|
self.client.post(full_uri)
|
||||||
}
|
}
|
||||||
|
|
||||||
|
pub async fn send(&self, req: RequestBuilder) -> Result<Response> {
|
||||||
|
self.sender.send(req).await
|
||||||
|
}
|
||||||
|
|
||||||
async fn rsp_to_str(response: Response) -> String {
|
async fn rsp_to_str(response: Response) -> String {
|
||||||
let status = response.status();
|
let status = response.status();
|
||||||
response.text().await.unwrap_or_else(|_| status.to_string())
|
response.text().await.unwrap_or_else(|_| status.to_string())
|
||||||
@@ -126,3 +153,49 @@ impl RestfulLanceDbClient {
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[cfg(test)]
|
||||||
|
pub mod test_utils {
|
||||||
|
use std::sync::Arc;
|
||||||
|
|
||||||
|
use super::*;
|
||||||
|
|
||||||
|
#[derive(Clone)]
|
||||||
|
pub struct MockSender {
|
||||||
|
f: Arc<dyn Fn(reqwest::Request) -> reqwest::Response + Send + Sync + 'static>,
|
||||||
|
}
|
||||||
|
|
||||||
|
impl std::fmt::Debug for MockSender {
|
||||||
|
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
||||||
|
write!(f, "MockSender")
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
impl HttpSend for MockSender {
|
||||||
|
async fn send(&self, request: reqwest::RequestBuilder) -> Result<reqwest::Response> {
|
||||||
|
let request = request.build().unwrap();
|
||||||
|
let response = (self.f)(request);
|
||||||
|
Ok(response)
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
pub fn client_with_handler<T>(
|
||||||
|
handler: impl Fn(reqwest::Request) -> http::response::Response<T> + Send + Sync + 'static,
|
||||||
|
) -> RestfulLanceDbClient<MockSender>
|
||||||
|
where
|
||||||
|
T: Into<reqwest::Body>,
|
||||||
|
{
|
||||||
|
let wrapper = move |req: reqwest::Request| {
|
||||||
|
let response = handler(req);
|
||||||
|
response.into()
|
||||||
|
};
|
||||||
|
|
||||||
|
RestfulLanceDbClient {
|
||||||
|
client: reqwest::Client::new(),
|
||||||
|
host: "http://localhost".to_string(),
|
||||||
|
sender: MockSender {
|
||||||
|
f: Arc::new(wrapper),
|
||||||
|
},
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|||||||
@@ -16,6 +16,7 @@ use std::sync::Arc;
|
|||||||
|
|
||||||
use arrow_array::RecordBatchReader;
|
use arrow_array::RecordBatchReader;
|
||||||
use async_trait::async_trait;
|
use async_trait::async_trait;
|
||||||
|
use http::StatusCode;
|
||||||
use reqwest::header::CONTENT_TYPE;
|
use reqwest::header::CONTENT_TYPE;
|
||||||
use serde::Deserialize;
|
use serde::Deserialize;
|
||||||
use tokio::task::spawn_blocking;
|
use tokio::task::spawn_blocking;
|
||||||
@@ -27,11 +28,10 @@ use crate::embeddings::EmbeddingRegistry;
|
|||||||
use crate::error::Result;
|
use crate::error::Result;
|
||||||
use crate::Table;
|
use crate::Table;
|
||||||
|
|
||||||
use super::client::RestfulLanceDbClient;
|
use super::client::{HttpSend, RestfulLanceDbClient, Sender};
|
||||||
use super::table::RemoteTable;
|
use super::table::RemoteTable;
|
||||||
use super::util::batches_to_ipc_bytes;
|
use super::util::batches_to_ipc_bytes;
|
||||||
|
use super::ARROW_STREAM_CONTENT_TYPE;
|
||||||
const ARROW_STREAM_CONTENT_TYPE: &str = "application/vnd.apache.arrow.stream";
|
|
||||||
|
|
||||||
#[derive(Deserialize)]
|
#[derive(Deserialize)]
|
||||||
struct ListTablesResponse {
|
struct ListTablesResponse {
|
||||||
@@ -39,8 +39,8 @@ struct ListTablesResponse {
|
|||||||
}
|
}
|
||||||
|
|
||||||
#[derive(Debug)]
|
#[derive(Debug)]
|
||||||
pub struct RemoteDatabase {
|
pub struct RemoteDatabase<S: HttpSend = Sender> {
|
||||||
client: RestfulLanceDbClient,
|
client: RestfulLanceDbClient<S>,
|
||||||
}
|
}
|
||||||
|
|
||||||
impl RemoteDatabase {
|
impl RemoteDatabase {
|
||||||
@@ -55,14 +55,32 @@ impl RemoteDatabase {
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
impl std::fmt::Display for RemoteDatabase {
|
#[cfg(all(test, feature = "remote"))]
|
||||||
|
mod test_utils {
|
||||||
|
use super::*;
|
||||||
|
use crate::remote::client::test_utils::client_with_handler;
|
||||||
|
use crate::remote::client::test_utils::MockSender;
|
||||||
|
|
||||||
|
impl RemoteDatabase<MockSender> {
|
||||||
|
pub fn new_mock<F, T>(handler: F) -> Self
|
||||||
|
where
|
||||||
|
F: Fn(reqwest::Request) -> http::Response<T> + Send + Sync + 'static,
|
||||||
|
T: Into<reqwest::Body>,
|
||||||
|
{
|
||||||
|
let client = client_with_handler(handler);
|
||||||
|
Self { client }
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
impl<S: HttpSend> std::fmt::Display for RemoteDatabase<S> {
|
||||||
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
||||||
write!(f, "RemoteDatabase(host={})", self.client.host())
|
write!(f, "RemoteDatabase(host={})", self.client.host())
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
#[async_trait]
|
#[async_trait]
|
||||||
impl ConnectionInternal for RemoteDatabase {
|
impl<S: HttpSend> ConnectionInternal for RemoteDatabase<S> {
|
||||||
async fn table_names(&self, options: TableNamesBuilder) -> Result<Vec<String>> {
|
async fn table_names(&self, options: TableNamesBuilder) -> Result<Vec<String>> {
|
||||||
let mut req = self.client.get("/v1/table/");
|
let mut req = self.client.get("/v1/table/");
|
||||||
if let Some(limit) = options.limit {
|
if let Some(limit) = options.limit {
|
||||||
@@ -71,7 +89,7 @@ impl ConnectionInternal for RemoteDatabase {
|
|||||||
if let Some(start_after) = options.start_after {
|
if let Some(start_after) = options.start_after {
|
||||||
req = req.query(&[("page_token", start_after)]);
|
req = req.query(&[("page_token", start_after)]);
|
||||||
}
|
}
|
||||||
let rsp = req.send().await?;
|
let rsp = self.client.send(req).await?;
|
||||||
let rsp = self.client.check_response(rsp).await?;
|
let rsp = self.client.check_response(rsp).await?;
|
||||||
Ok(rsp.json::<ListTablesResponse>().await?.tables)
|
Ok(rsp.json::<ListTablesResponse>().await?.tables)
|
||||||
}
|
}
|
||||||
@@ -88,15 +106,24 @@ impl ConnectionInternal for RemoteDatabase {
|
|||||||
.await
|
.await
|
||||||
.unwrap()?;
|
.unwrap()?;
|
||||||
|
|
||||||
let rsp = self
|
let req = self
|
||||||
.client
|
.client
|
||||||
.post(&format!("/v1/table/{}/create/", options.name))
|
.post(&format!("/v1/table/{}/create/", options.name))
|
||||||
.body(data_buffer)
|
.body(data_buffer)
|
||||||
.header(CONTENT_TYPE, ARROW_STREAM_CONTENT_TYPE)
|
.header(CONTENT_TYPE, ARROW_STREAM_CONTENT_TYPE)
|
||||||
// This is currently expected by LanceDb cloud but will be removed soon.
|
// This is currently expected by LanceDb cloud but will be removed soon.
|
||||||
.header("x-request-id", "na")
|
.header("x-request-id", "na");
|
||||||
.send()
|
let rsp = self.client.send(req).await?;
|
||||||
.await?;
|
|
||||||
|
if rsp.status() == StatusCode::BAD_REQUEST {
|
||||||
|
let body = rsp.text().await?;
|
||||||
|
if body.contains("already exists") {
|
||||||
|
return Err(crate::Error::TableAlreadyExists { name: options.name });
|
||||||
|
} else {
|
||||||
|
return Err(crate::Error::InvalidInput { message: body });
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
self.client.check_response(rsp).await?;
|
self.client.check_response(rsp).await?;
|
||||||
|
|
||||||
Ok(Table::new(Arc::new(RemoteTable::new(
|
Ok(Table::new(Arc::new(RemoteTable::new(
|
||||||
@@ -105,19 +132,206 @@ impl ConnectionInternal for RemoteDatabase {
|
|||||||
))))
|
))))
|
||||||
}
|
}
|
||||||
|
|
||||||
async fn do_open_table(&self, _options: OpenTableBuilder) -> Result<Table> {
|
async fn do_open_table(&self, options: OpenTableBuilder) -> Result<Table> {
|
||||||
todo!()
|
// We describe the table to confirm it exists before moving on.
|
||||||
|
// TODO: a TTL cache of table existence
|
||||||
|
let req = self
|
||||||
|
.client
|
||||||
|
.get(&format!("/v1/table/{}/describe/", options.name));
|
||||||
|
let resp = self.client.send(req).await?;
|
||||||
|
if resp.status() == StatusCode::NOT_FOUND {
|
||||||
|
return Err(crate::Error::TableNotFound { name: options.name });
|
||||||
|
}
|
||||||
|
self.client.check_response(resp).await?;
|
||||||
|
Ok(Table::new(Arc::new(RemoteTable::new(
|
||||||
|
self.client.clone(),
|
||||||
|
options.name,
|
||||||
|
))))
|
||||||
}
|
}
|
||||||
|
|
||||||
async fn drop_table(&self, _name: &str) -> Result<()> {
|
async fn drop_table(&self, name: &str) -> Result<()> {
|
||||||
todo!()
|
let req = self.client.post(&format!("/v1/table/{}/drop/", name));
|
||||||
|
let resp = self.client.send(req).await?;
|
||||||
|
self.client.check_response(resp).await?;
|
||||||
|
Ok(())
|
||||||
}
|
}
|
||||||
|
|
||||||
async fn drop_db(&self) -> Result<()> {
|
async fn drop_db(&self) -> Result<()> {
|
||||||
todo!()
|
Err(crate::Error::NotSupported {
|
||||||
|
message: "Dropping databases is not supported in the remote API".to_string(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
|
|
||||||
fn embedding_registry(&self) -> &dyn EmbeddingRegistry {
|
fn embedding_registry(&self) -> &dyn EmbeddingRegistry {
|
||||||
todo!()
|
todo!()
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[cfg(test)]
|
||||||
|
mod tests {
|
||||||
|
use std::sync::Arc;
|
||||||
|
|
||||||
|
use arrow_array::{Int32Array, RecordBatch, RecordBatchIterator};
|
||||||
|
use arrow_schema::{DataType, Field, Schema};
|
||||||
|
|
||||||
|
use crate::{remote::db::ARROW_STREAM_CONTENT_TYPE, Connection};
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_table_names() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::GET);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/");
|
||||||
|
assert_eq!(request.url().query(), None);
|
||||||
|
|
||||||
|
http::Response::builder()
|
||||||
|
.status(200)
|
||||||
|
.body(r#"{"tables": ["table1", "table2"]}"#)
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
let names = conn.table_names().execute().await.unwrap();
|
||||||
|
assert_eq!(names, vec!["table1", "table2"]);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_table_names_pagination() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::GET);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/");
|
||||||
|
assert!(request.url().query().unwrap().contains("limit=2"));
|
||||||
|
assert!(request.url().query().unwrap().contains("page_token=table2"));
|
||||||
|
|
||||||
|
http::Response::builder()
|
||||||
|
.status(200)
|
||||||
|
.body(r#"{"tables": ["table3", "table4"], "page_token": "token"}"#)
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
let names = conn
|
||||||
|
.table_names()
|
||||||
|
.start_after("table2")
|
||||||
|
.limit(2)
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
assert_eq!(names, vec!["table3", "table4"]);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_open_table() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::GET);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/table1/describe/");
|
||||||
|
assert_eq!(request.url().query(), None);
|
||||||
|
|
||||||
|
http::Response::builder()
|
||||||
|
.status(200)
|
||||||
|
.body(r#"{"table": "table1"}"#)
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
let table = conn.open_table("table1").execute().await.unwrap();
|
||||||
|
assert_eq!(table.name(), "table1");
|
||||||
|
|
||||||
|
// Storage options should be ignored.
|
||||||
|
let table = conn
|
||||||
|
.open_table("table1")
|
||||||
|
.storage_option("key", "value")
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
assert_eq!(table.name(), "table1");
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_open_table_not_found() {
|
||||||
|
let conn = Connection::new_with_handler(|_| {
|
||||||
|
http::Response::builder()
|
||||||
|
.status(404)
|
||||||
|
.body("table not found")
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
let result = conn.open_table("table1").execute().await;
|
||||||
|
assert!(result.is_err());
|
||||||
|
assert!(matches!(result, Err(crate::Error::TableNotFound { .. })));
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_create_table() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::POST);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/table1/create/");
|
||||||
|
assert_eq!(
|
||||||
|
request
|
||||||
|
.headers()
|
||||||
|
.get(reqwest::header::CONTENT_TYPE)
|
||||||
|
.unwrap(),
|
||||||
|
ARROW_STREAM_CONTENT_TYPE.as_bytes()
|
||||||
|
);
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
let data = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
let reader = RecordBatchIterator::new([Ok(data.clone())], data.schema());
|
||||||
|
let table = conn.create_table("table1", reader).execute().await.unwrap();
|
||||||
|
assert_eq!(table.name(), "table1");
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_create_table_already_exists() {
|
||||||
|
let conn = Connection::new_with_handler(|_| {
|
||||||
|
http::Response::builder()
|
||||||
|
.status(400)
|
||||||
|
.body("table table1 already exists")
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
let data = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
let reader = RecordBatchIterator::new([Ok(data.clone())], data.schema());
|
||||||
|
let result = conn.create_table("table1", reader).execute().await;
|
||||||
|
assert!(result.is_err());
|
||||||
|
assert!(
|
||||||
|
matches!(result, Err(crate::Error::TableAlreadyExists { name }) if name == "table1")
|
||||||
|
);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_create_table_empty() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::POST);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/table1/create/");
|
||||||
|
assert_eq!(
|
||||||
|
request
|
||||||
|
.headers()
|
||||||
|
.get(reqwest::header::CONTENT_TYPE)
|
||||||
|
.unwrap(),
|
||||||
|
ARROW_STREAM_CONTENT_TYPE.as_bytes()
|
||||||
|
);
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
let schema = Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)]));
|
||||||
|
conn.create_empty_table("table1", schema)
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_drop_table() {
|
||||||
|
let conn = Connection::new_with_handler(|request| {
|
||||||
|
assert_eq!(request.method(), &reqwest::Method::POST);
|
||||||
|
assert_eq!(request.url().path(), "/v1/table/table1/drop/");
|
||||||
|
assert_eq!(request.url().query(), None);
|
||||||
|
assert!(request.body().is_none());
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
conn.drop_table("table1").await.unwrap();
|
||||||
|
// NOTE: the API will return 200 even if the table does not exist. So we shouldn't expect 404.
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|||||||
@@ -1,12 +1,18 @@
|
|||||||
use std::sync::Arc;
|
use std::sync::{Arc, Mutex};
|
||||||
|
|
||||||
use crate::table::dataset::DatasetReadGuard;
|
use crate::table::dataset::DatasetReadGuard;
|
||||||
|
use crate::table::AddDataMode;
|
||||||
|
use crate::Error;
|
||||||
use arrow_array::RecordBatchReader;
|
use arrow_array::RecordBatchReader;
|
||||||
use arrow_schema::SchemaRef;
|
use arrow_schema::SchemaRef;
|
||||||
use async_trait::async_trait;
|
use async_trait::async_trait;
|
||||||
use datafusion_physical_plan::ExecutionPlan;
|
use datafusion_physical_plan::ExecutionPlan;
|
||||||
|
use http::header::CONTENT_TYPE;
|
||||||
|
use http::StatusCode;
|
||||||
|
use lance::arrow::json::JsonSchema;
|
||||||
use lance::dataset::scanner::{DatasetRecordBatchStream, Scanner};
|
use lance::dataset::scanner::{DatasetRecordBatchStream, Scanner};
|
||||||
use lance::dataset::{ColumnAlteration, NewColumnTransform};
|
use lance::dataset::{ColumnAlteration, NewColumnTransform};
|
||||||
|
use serde::{Deserialize, Serialize};
|
||||||
|
|
||||||
use crate::{
|
use crate::{
|
||||||
connection::NoData,
|
connection::NoData,
|
||||||
@@ -19,29 +25,100 @@ use crate::{
|
|||||||
},
|
},
|
||||||
};
|
};
|
||||||
|
|
||||||
use super::client::RestfulLanceDbClient;
|
use super::client::{HttpSend, RestfulLanceDbClient, Sender};
|
||||||
|
use super::ARROW_STREAM_CONTENT_TYPE;
|
||||||
|
|
||||||
#[derive(Debug)]
|
#[derive(Debug)]
|
||||||
pub struct RemoteTable {
|
pub struct RemoteTable<S: HttpSend = Sender> {
|
||||||
#[allow(dead_code)]
|
#[allow(dead_code)]
|
||||||
client: RestfulLanceDbClient,
|
client: RestfulLanceDbClient<S>,
|
||||||
name: String,
|
name: String,
|
||||||
}
|
}
|
||||||
|
|
||||||
impl RemoteTable {
|
impl<S: HttpSend> RemoteTable<S> {
|
||||||
pub fn new(client: RestfulLanceDbClient, name: String) -> Self {
|
pub fn new(client: RestfulLanceDbClient<S>, name: String) -> Self {
|
||||||
Self { client, name }
|
Self { client, name }
|
||||||
}
|
}
|
||||||
|
|
||||||
|
async fn describe(&self) -> Result<TableDescription> {
|
||||||
|
let request = self.client.post(&format!("/table/{}/describe/", self.name));
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
|
||||||
|
let response = self.check_table_response(response).await?;
|
||||||
|
|
||||||
|
let body = response.text().await?;
|
||||||
|
|
||||||
|
serde_json::from_str(&body).map_err(|e| Error::Http {
|
||||||
|
message: format!("Failed to parse table description: {}", e),
|
||||||
|
})
|
||||||
|
}
|
||||||
|
|
||||||
|
fn reader_as_body(data: Box<dyn RecordBatchReader + Send>) -> Result<reqwest::Body> {
|
||||||
|
// TODO: Once Phalanx supports compression, we should use it here.
|
||||||
|
let mut writer = arrow_ipc::writer::StreamWriter::try_new(Vec::new(), &data.schema())?;
|
||||||
|
|
||||||
|
// Mutex is just here to make it sync. We shouldn't have any contention.
|
||||||
|
let mut data = Mutex::new(data);
|
||||||
|
let body_iter = std::iter::from_fn(move || match data.get_mut().unwrap().next() {
|
||||||
|
Some(Ok(batch)) => {
|
||||||
|
writer.write(&batch).ok()?;
|
||||||
|
let buffer = std::mem::take(writer.get_mut());
|
||||||
|
Some(Ok(buffer))
|
||||||
|
}
|
||||||
|
Some(Err(e)) => Some(Err(e)),
|
||||||
|
None => {
|
||||||
|
writer.finish().ok()?;
|
||||||
|
let buffer = std::mem::take(writer.get_mut());
|
||||||
|
Some(Ok(buffer))
|
||||||
|
}
|
||||||
|
});
|
||||||
|
let body_stream = futures::stream::iter(body_iter);
|
||||||
|
Ok(reqwest::Body::wrap_stream(body_stream))
|
||||||
|
}
|
||||||
|
|
||||||
|
async fn check_table_response(&self, response: reqwest::Response) -> Result<reqwest::Response> {
|
||||||
|
if response.status() == StatusCode::NOT_FOUND {
|
||||||
|
return Err(Error::TableNotFound {
|
||||||
|
name: self.name.clone(),
|
||||||
|
});
|
||||||
|
}
|
||||||
|
|
||||||
|
self.client.check_response(response).await
|
||||||
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
impl std::fmt::Display for RemoteTable {
|
#[derive(Deserialize)]
|
||||||
|
struct TableDescription {
|
||||||
|
version: u64,
|
||||||
|
schema: JsonSchema,
|
||||||
|
}
|
||||||
|
|
||||||
|
impl<S: HttpSend> std::fmt::Display for RemoteTable<S> {
|
||||||
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
||||||
write!(f, "RemoteTable({})", self.name)
|
write!(f, "RemoteTable({})", self.name)
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[cfg(all(test, feature = "remote"))]
|
||||||
|
mod test_utils {
|
||||||
|
use super::*;
|
||||||
|
use crate::remote::client::test_utils::client_with_handler;
|
||||||
|
use crate::remote::client::test_utils::MockSender;
|
||||||
|
|
||||||
|
impl RemoteTable<MockSender> {
|
||||||
|
pub fn new_mock<F, T>(name: String, handler: F) -> Self
|
||||||
|
where
|
||||||
|
F: Fn(reqwest::Request) -> http::Response<T> + Send + Sync + 'static,
|
||||||
|
T: Into<reqwest::Body>,
|
||||||
|
{
|
||||||
|
let client = client_with_handler(handler);
|
||||||
|
Self { client, name }
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
#[async_trait]
|
#[async_trait]
|
||||||
impl TableInternal for RemoteTable {
|
impl<S: HttpSend> TableInternal for RemoteTable<S> {
|
||||||
fn as_any(&self) -> &dyn std::any::Any {
|
fn as_any(&self) -> &dyn std::any::Any {
|
||||||
self
|
self
|
||||||
}
|
}
|
||||||
@@ -52,29 +129,72 @@ impl TableInternal for RemoteTable {
|
|||||||
&self.name
|
&self.name
|
||||||
}
|
}
|
||||||
async fn version(&self) -> Result<u64> {
|
async fn version(&self) -> Result<u64> {
|
||||||
todo!()
|
self.describe().await.map(|desc| desc.version)
|
||||||
}
|
}
|
||||||
async fn checkout(&self, _version: u64) -> Result<()> {
|
async fn checkout(&self, _version: u64) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "checkout is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn checkout_latest(&self) -> Result<()> {
|
async fn checkout_latest(&self) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "checkout is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn restore(&self) -> Result<()> {
|
async fn restore(&self) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "restore is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn schema(&self) -> Result<SchemaRef> {
|
async fn schema(&self) -> Result<SchemaRef> {
|
||||||
todo!()
|
let schema = self.describe().await?.schema;
|
||||||
|
Ok(Arc::new(schema.try_into()?))
|
||||||
}
|
}
|
||||||
async fn count_rows(&self, _filter: Option<String>) -> Result<usize> {
|
async fn count_rows(&self, filter: Option<String>) -> Result<usize> {
|
||||||
todo!()
|
let mut request = self
|
||||||
|
.client
|
||||||
|
.post(&format!("/table/{}/count_rows/", self.name));
|
||||||
|
|
||||||
|
if let Some(filter) = filter {
|
||||||
|
request = request.json(&serde_json::json!({ "filter": filter }));
|
||||||
|
} else {
|
||||||
|
request = request.json(&serde_json::json!({}));
|
||||||
|
}
|
||||||
|
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
|
||||||
|
let response = self.check_table_response(response).await?;
|
||||||
|
|
||||||
|
let body = response.text().await?;
|
||||||
|
|
||||||
|
serde_json::from_str(&body).map_err(|e| Error::Http {
|
||||||
|
message: format!("Failed to parse row count: {}", e),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn add(
|
async fn add(
|
||||||
&self,
|
&self,
|
||||||
_add: AddDataBuilder<NoData>,
|
add: AddDataBuilder<NoData>,
|
||||||
_data: Box<dyn RecordBatchReader + Send>,
|
data: Box<dyn RecordBatchReader + Send>,
|
||||||
) -> Result<()> {
|
) -> Result<()> {
|
||||||
todo!()
|
let body = Self::reader_as_body(data)?;
|
||||||
|
let mut request = self
|
||||||
|
.client
|
||||||
|
.post(&format!("/table/{}/insert/", self.name))
|
||||||
|
.header(CONTENT_TYPE, ARROW_STREAM_CONTENT_TYPE)
|
||||||
|
.body(body);
|
||||||
|
|
||||||
|
match add.mode {
|
||||||
|
AddDataMode::Append => {}
|
||||||
|
AddDataMode::Overwrite => {
|
||||||
|
request = request.query(&[("mode", "overwrite")]);
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
|
||||||
|
self.check_table_response(response).await?;
|
||||||
|
|
||||||
|
Ok(())
|
||||||
}
|
}
|
||||||
async fn build_plan(
|
async fn build_plan(
|
||||||
&self,
|
&self,
|
||||||
@@ -82,61 +202,530 @@ impl TableInternal for RemoteTable {
|
|||||||
_query: &VectorQuery,
|
_query: &VectorQuery,
|
||||||
_options: Option<QueryExecutionOptions>,
|
_options: Option<QueryExecutionOptions>,
|
||||||
) -> Result<Scanner> {
|
) -> Result<Scanner> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "build_plan is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn create_plan(
|
async fn create_plan(
|
||||||
&self,
|
&self,
|
||||||
_query: &VectorQuery,
|
_query: &VectorQuery,
|
||||||
_options: QueryExecutionOptions,
|
_options: QueryExecutionOptions,
|
||||||
) -> Result<Arc<dyn ExecutionPlan>> {
|
) -> Result<Arc<dyn ExecutionPlan>> {
|
||||||
unimplemented!()
|
Err(Error::NotSupported {
|
||||||
|
message: "create_plan is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn explain_plan(&self, _query: &VectorQuery, _verbose: bool) -> Result<String> {
|
async fn explain_plan(&self, _query: &VectorQuery, _verbose: bool) -> Result<String> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "explain_plan is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn plain_query(
|
async fn plain_query(
|
||||||
&self,
|
&self,
|
||||||
_query: &Query,
|
_query: &Query,
|
||||||
_options: QueryExecutionOptions,
|
_options: QueryExecutionOptions,
|
||||||
) -> Result<DatasetRecordBatchStream> {
|
) -> Result<DatasetRecordBatchStream> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "plain_query is not yet supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn update(&self, _update: UpdateBuilder) -> Result<()> {
|
async fn update(&self, update: UpdateBuilder) -> Result<u64> {
|
||||||
todo!()
|
let request = self.client.post(&format!("/table/{}/update/", self.name));
|
||||||
|
|
||||||
|
let mut updates = Vec::new();
|
||||||
|
for (column, expression) in update.columns {
|
||||||
|
updates.push(column);
|
||||||
|
updates.push(expression);
|
||||||
|
}
|
||||||
|
|
||||||
|
let request = request.json(&serde_json::json!({
|
||||||
|
"updates": updates,
|
||||||
|
"only_if": update.filter,
|
||||||
|
}));
|
||||||
|
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
|
||||||
|
let response = self.check_table_response(response).await?;
|
||||||
|
|
||||||
|
let body = response.text().await?;
|
||||||
|
|
||||||
|
serde_json::from_str(&body).map_err(|e| Error::Http {
|
||||||
|
message: format!(
|
||||||
|
"Failed to parse updated rows result from response {}: {}",
|
||||||
|
body, e
|
||||||
|
),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn delete(&self, _predicate: &str) -> Result<()> {
|
async fn delete(&self, predicate: &str) -> Result<()> {
|
||||||
todo!()
|
let body = serde_json::json!({ "predicate": predicate });
|
||||||
|
let request = self
|
||||||
|
.client
|
||||||
|
.post(&format!("/table/{}/delete/", self.name))
|
||||||
|
.json(&body);
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
self.check_table_response(response).await?;
|
||||||
|
Ok(())
|
||||||
}
|
}
|
||||||
async fn create_index(&self, _index: IndexBuilder) -> Result<()> {
|
async fn create_index(&self, _index: IndexBuilder) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "create_index is not yet supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn merge_insert(
|
async fn merge_insert(
|
||||||
&self,
|
&self,
|
||||||
_params: MergeInsertBuilder,
|
params: MergeInsertBuilder,
|
||||||
_new_data: Box<dyn RecordBatchReader + Send>,
|
new_data: Box<dyn RecordBatchReader + Send>,
|
||||||
) -> Result<()> {
|
) -> Result<()> {
|
||||||
todo!()
|
let query = MergeInsertRequest::try_from(params)?;
|
||||||
|
let body = Self::reader_as_body(new_data)?;
|
||||||
|
let request = self
|
||||||
|
.client
|
||||||
|
.post(&format!("/table/{}/merge_insert/", self.name))
|
||||||
|
.query(&query)
|
||||||
|
.header(CONTENT_TYPE, ARROW_STREAM_CONTENT_TYPE)
|
||||||
|
.body(body);
|
||||||
|
|
||||||
|
let response = self.client.send(request).await?;
|
||||||
|
|
||||||
|
self.check_table_response(response).await?;
|
||||||
|
|
||||||
|
Ok(())
|
||||||
}
|
}
|
||||||
async fn optimize(&self, _action: OptimizeAction) -> Result<OptimizeStats> {
|
async fn optimize(&self, _action: OptimizeAction) -> Result<OptimizeStats> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "optimize is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn add_columns(
|
async fn add_columns(
|
||||||
&self,
|
&self,
|
||||||
_transforms: NewColumnTransform,
|
_transforms: NewColumnTransform,
|
||||||
_read_columns: Option<Vec<String>>,
|
_read_columns: Option<Vec<String>>,
|
||||||
) -> Result<()> {
|
) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "add_columns is not yet supported.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn alter_columns(&self, _alterations: &[ColumnAlteration]) -> Result<()> {
|
async fn alter_columns(&self, _alterations: &[ColumnAlteration]) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "alter_columns is not yet supported.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn drop_columns(&self, _columns: &[&str]) -> Result<()> {
|
async fn drop_columns(&self, _columns: &[&str]) -> Result<()> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "drop_columns is not yet supported.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn list_indices(&self) -> Result<Vec<IndexConfig>> {
|
async fn list_indices(&self) -> Result<Vec<IndexConfig>> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "list_indices is not yet supported.".into(),
|
||||||
|
})
|
||||||
}
|
}
|
||||||
async fn table_definition(&self) -> Result<TableDefinition> {
|
async fn table_definition(&self) -> Result<TableDefinition> {
|
||||||
todo!()
|
Err(Error::NotSupported {
|
||||||
|
message: "table_definition is not supported on LanceDB cloud.".into(),
|
||||||
|
})
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#[derive(Serialize)]
|
||||||
|
struct MergeInsertRequest {
|
||||||
|
on: String,
|
||||||
|
when_matched_update_all: bool,
|
||||||
|
when_matched_update_all_filt: Option<String>,
|
||||||
|
when_not_matched_insert_all: bool,
|
||||||
|
when_not_matched_by_source_delete: bool,
|
||||||
|
when_not_matched_by_source_delete_filt: Option<String>,
|
||||||
|
}
|
||||||
|
|
||||||
|
impl TryFrom<MergeInsertBuilder> for MergeInsertRequest {
|
||||||
|
type Error = Error;
|
||||||
|
|
||||||
|
fn try_from(value: MergeInsertBuilder) -> Result<Self> {
|
||||||
|
if value.on.is_empty() {
|
||||||
|
return Err(Error::InvalidInput {
|
||||||
|
message: "MergeInsertBuilder missing required 'on' field".into(),
|
||||||
|
});
|
||||||
|
} else if value.on.len() > 1 {
|
||||||
|
return Err(Error::NotSupported {
|
||||||
|
message: "MergeInsertBuilder only supports a single 'on' column".into(),
|
||||||
|
});
|
||||||
|
}
|
||||||
|
let on = value.on[0].clone();
|
||||||
|
|
||||||
|
Ok(Self {
|
||||||
|
on,
|
||||||
|
when_matched_update_all: value.when_matched_update_all,
|
||||||
|
when_matched_update_all_filt: value.when_matched_update_all_filt,
|
||||||
|
when_not_matched_insert_all: value.when_not_matched_insert_all,
|
||||||
|
when_not_matched_by_source_delete: value.when_not_matched_by_source_delete,
|
||||||
|
when_not_matched_by_source_delete_filt: value.when_not_matched_by_source_delete_filt,
|
||||||
|
})
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#[cfg(test)]
|
||||||
|
mod tests {
|
||||||
|
use std::{collections::HashMap, pin::Pin};
|
||||||
|
|
||||||
|
use super::*;
|
||||||
|
|
||||||
|
use arrow_array::{Int32Array, RecordBatch, RecordBatchIterator};
|
||||||
|
use arrow_schema::{DataType, Field, Schema};
|
||||||
|
use futures::{future::BoxFuture, StreamExt, TryFutureExt};
|
||||||
|
use reqwest::Body;
|
||||||
|
|
||||||
|
use crate::{Error, Table};
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_not_found() {
|
||||||
|
let table = Table::new_with_handler("my_table", |_| {
|
||||||
|
http::Response::builder()
|
||||||
|
.status(404)
|
||||||
|
.body("table my_table not found")
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
let batch = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
let example_data = || {
|
||||||
|
Box::new(RecordBatchIterator::new(
|
||||||
|
[Ok(batch.clone())],
|
||||||
|
batch.schema(),
|
||||||
|
))
|
||||||
|
};
|
||||||
|
|
||||||
|
// All endpoints should translate 404 to TableNotFound.
|
||||||
|
let results: Vec<BoxFuture<'_, Result<()>>> = vec![
|
||||||
|
Box::pin(table.version().map_ok(|_| ())),
|
||||||
|
Box::pin(table.schema().map_ok(|_| ())),
|
||||||
|
Box::pin(table.count_rows(None).map_ok(|_| ())),
|
||||||
|
Box::pin(table.update().column("a", "a + 1").execute().map_ok(|_| ())),
|
||||||
|
Box::pin(table.add(example_data()).execute().map_ok(|_| ())),
|
||||||
|
Box::pin(table.merge_insert(&["test"]).execute(example_data())),
|
||||||
|
Box::pin(table.delete("false")), // TODO: other endpoints.
|
||||||
|
];
|
||||||
|
|
||||||
|
for result in results {
|
||||||
|
let result = result.await;
|
||||||
|
assert!(result.is_err());
|
||||||
|
assert!(matches!(result, Err(Error::TableNotFound { name }) if name == "my_table"));
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_version() {
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/describe/");
|
||||||
|
|
||||||
|
http::Response::builder()
|
||||||
|
.status(200)
|
||||||
|
.body(r#"{"version": 42, "schema": { "fields": [] }}"#)
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
let version = table.version().await.unwrap();
|
||||||
|
assert_eq!(version, 42);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_schema() {
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/describe/");
|
||||||
|
|
||||||
|
http::Response::builder()
|
||||||
|
.status(200)
|
||||||
|
.body(
|
||||||
|
r#"{"version": 42, "schema": {"fields": [
|
||||||
|
{"name": "a", "type": { "type": "int32" }, "nullable": false},
|
||||||
|
{"name": "b", "type": { "type": "string" }, "nullable": true}
|
||||||
|
], "metadata": {"key": "value"}}}"#,
|
||||||
|
)
|
||||||
|
.unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
let expected = Arc::new(
|
||||||
|
Schema::new(vec![
|
||||||
|
Field::new("a", DataType::Int32, false),
|
||||||
|
Field::new("b", DataType::Utf8, true),
|
||||||
|
])
|
||||||
|
.with_metadata([("key".into(), "value".into())].into()),
|
||||||
|
);
|
||||||
|
|
||||||
|
let schema = table.schema().await.unwrap();
|
||||||
|
assert_eq!(schema, expected);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_count_rows() {
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/count_rows/");
|
||||||
|
assert_eq!(request.body().unwrap().as_bytes().unwrap(), br#"{}"#);
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("42").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
let count = table.count_rows(None).await.unwrap();
|
||||||
|
assert_eq!(count, 42);
|
||||||
|
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/count_rows/");
|
||||||
|
assert_eq!(
|
||||||
|
request.body().unwrap().as_bytes().unwrap(),
|
||||||
|
br#"{"filter":"a > 10"}"#
|
||||||
|
);
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("42").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
let count = table.count_rows(Some("a > 10".into())).await.unwrap();
|
||||||
|
assert_eq!(count, 42);
|
||||||
|
}
|
||||||
|
|
||||||
|
async fn collect_body(body: Body) -> Vec<u8> {
|
||||||
|
use http_body::Body;
|
||||||
|
let mut body = body;
|
||||||
|
let mut data = Vec::new();
|
||||||
|
let mut body_pin = Pin::new(&mut body);
|
||||||
|
futures::stream::poll_fn(|cx| body_pin.as_mut().poll_frame(cx))
|
||||||
|
.for_each(|frame| {
|
||||||
|
data.extend_from_slice(frame.unwrap().data_ref().unwrap());
|
||||||
|
futures::future::ready(())
|
||||||
|
})
|
||||||
|
.await;
|
||||||
|
data
|
||||||
|
}
|
||||||
|
|
||||||
|
fn write_ipc_stream(data: &RecordBatch) -> Vec<u8> {
|
||||||
|
let mut body = Vec::new();
|
||||||
|
{
|
||||||
|
let mut writer = arrow_ipc::writer::StreamWriter::try_new(&mut body, &data.schema())
|
||||||
|
.expect("Failed to create writer");
|
||||||
|
writer.write(data).expect("Failed to write data");
|
||||||
|
writer.finish().expect("Failed to finish");
|
||||||
|
}
|
||||||
|
body
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_add_append() {
|
||||||
|
let data = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
|
||||||
|
let (sender, receiver) = std::sync::mpsc::channel();
|
||||||
|
let table = Table::new_with_handler("my_table", move |mut request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/insert/");
|
||||||
|
// If mode is specified, it should be "append". Append is default
|
||||||
|
// so it's not required.
|
||||||
|
assert!(request
|
||||||
|
.url()
|
||||||
|
.query_pairs()
|
||||||
|
.filter(|(k, _)| k == "mode")
|
||||||
|
.all(|(_, v)| v == "append"));
|
||||||
|
|
||||||
|
assert_eq!(
|
||||||
|
request.headers().get("Content-Type").unwrap(),
|
||||||
|
ARROW_STREAM_CONTENT_TYPE
|
||||||
|
);
|
||||||
|
|
||||||
|
let mut body_out = reqwest::Body::from(Vec::new());
|
||||||
|
std::mem::swap(request.body_mut().as_mut().unwrap(), &mut body_out);
|
||||||
|
sender.send(body_out).unwrap();
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
table
|
||||||
|
.add(RecordBatchIterator::new([Ok(data.clone())], data.schema()))
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
|
||||||
|
let body = receiver.recv().unwrap();
|
||||||
|
let body = collect_body(body).await;
|
||||||
|
let expected_body = write_ipc_stream(&data);
|
||||||
|
assert_eq!(&body, &expected_body);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_add_overwrite() {
|
||||||
|
let data = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
|
||||||
|
let (sender, receiver) = std::sync::mpsc::channel();
|
||||||
|
let table = Table::new_with_handler("my_table", move |mut request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/insert/");
|
||||||
|
assert_eq!(
|
||||||
|
request
|
||||||
|
.url()
|
||||||
|
.query_pairs()
|
||||||
|
.find(|(k, _)| k == "mode")
|
||||||
|
.map(|kv| kv.1)
|
||||||
|
.as_deref(),
|
||||||
|
Some("overwrite"),
|
||||||
|
"Expected mode=overwrite"
|
||||||
|
);
|
||||||
|
|
||||||
|
assert_eq!(
|
||||||
|
request.headers().get("Content-Type").unwrap(),
|
||||||
|
ARROW_STREAM_CONTENT_TYPE
|
||||||
|
);
|
||||||
|
|
||||||
|
let mut body_out = reqwest::Body::from(Vec::new());
|
||||||
|
std::mem::swap(request.body_mut().as_mut().unwrap(), &mut body_out);
|
||||||
|
sender.send(body_out).unwrap();
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
table
|
||||||
|
.add(RecordBatchIterator::new([Ok(data.clone())], data.schema()))
|
||||||
|
.mode(AddDataMode::Overwrite)
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
|
||||||
|
let body = receiver.recv().unwrap();
|
||||||
|
let body = collect_body(body).await;
|
||||||
|
let expected_body = write_ipc_stream(&data);
|
||||||
|
assert_eq!(&body, &expected_body);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_update() {
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/update/");
|
||||||
|
|
||||||
|
if let Some(body) = request.body().unwrap().as_bytes() {
|
||||||
|
let body = std::str::from_utf8(body).unwrap();
|
||||||
|
let value: serde_json::Value = serde_json::from_str(body).unwrap();
|
||||||
|
let updates = value.get("updates").unwrap().as_array().unwrap();
|
||||||
|
assert!(updates.len() == 2);
|
||||||
|
|
||||||
|
let col_name = updates[0].as_str().unwrap();
|
||||||
|
let expression = updates[1].as_str().unwrap();
|
||||||
|
assert_eq!(col_name, "a");
|
||||||
|
assert_eq!(expression, "a + 1");
|
||||||
|
|
||||||
|
let only_if = value.get("only_if").unwrap().as_str().unwrap();
|
||||||
|
assert_eq!(only_if, "b > 10");
|
||||||
|
}
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("1").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
table
|
||||||
|
.update()
|
||||||
|
.column("a", "a + 1")
|
||||||
|
.only_if("b > 10")
|
||||||
|
.execute()
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_merge_insert() {
|
||||||
|
let batch = RecordBatch::try_new(
|
||||||
|
Arc::new(Schema::new(vec![Field::new("a", DataType::Int32, false)])),
|
||||||
|
vec![Arc::new(Int32Array::from(vec![1, 2, 3]))],
|
||||||
|
)
|
||||||
|
.unwrap();
|
||||||
|
let data = Box::new(RecordBatchIterator::new(
|
||||||
|
[Ok(batch.clone())],
|
||||||
|
batch.schema(),
|
||||||
|
));
|
||||||
|
|
||||||
|
// Default parameters
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/merge_insert/");
|
||||||
|
|
||||||
|
let params = request.url().query_pairs().collect::<HashMap<_, _>>();
|
||||||
|
assert_eq!(params["on"], "some_col");
|
||||||
|
assert_eq!(params["when_matched_update_all"], "false");
|
||||||
|
assert_eq!(params["when_not_matched_insert_all"], "false");
|
||||||
|
assert_eq!(params["when_not_matched_by_source_delete"], "false");
|
||||||
|
assert!(!params.contains_key("when_matched_update_all_filt"));
|
||||||
|
assert!(!params.contains_key("when_not_matched_by_source_delete_filt"));
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
table
|
||||||
|
.merge_insert(&["some_col"])
|
||||||
|
.execute(data)
|
||||||
|
.await
|
||||||
|
.unwrap();
|
||||||
|
|
||||||
|
// All parameters specified
|
||||||
|
let (sender, receiver) = std::sync::mpsc::channel();
|
||||||
|
let table = Table::new_with_handler("my_table", move |mut request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/merge_insert/");
|
||||||
|
assert_eq!(
|
||||||
|
request.headers().get("Content-Type").unwrap(),
|
||||||
|
ARROW_STREAM_CONTENT_TYPE
|
||||||
|
);
|
||||||
|
|
||||||
|
let params = request.url().query_pairs().collect::<HashMap<_, _>>();
|
||||||
|
assert_eq!(params["on"], "some_col");
|
||||||
|
assert_eq!(params["when_matched_update_all"], "true");
|
||||||
|
assert_eq!(params["when_not_matched_insert_all"], "false");
|
||||||
|
assert_eq!(params["when_not_matched_by_source_delete"], "true");
|
||||||
|
assert_eq!(params["when_matched_update_all_filt"], "a = 1");
|
||||||
|
assert_eq!(params["when_not_matched_by_source_delete_filt"], "b = 2");
|
||||||
|
|
||||||
|
let mut body_out = reqwest::Body::from(Vec::new());
|
||||||
|
std::mem::swap(request.body_mut().as_mut().unwrap(), &mut body_out);
|
||||||
|
sender.send(body_out).unwrap();
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
let mut builder = table.merge_insert(&["some_col"]);
|
||||||
|
builder
|
||||||
|
.when_matched_update_all(Some("a = 1".into()))
|
||||||
|
.when_not_matched_by_source_delete(Some("b = 2".into()));
|
||||||
|
let data = Box::new(RecordBatchIterator::new(
|
||||||
|
[Ok(batch.clone())],
|
||||||
|
batch.schema(),
|
||||||
|
));
|
||||||
|
builder.execute(data).await.unwrap();
|
||||||
|
|
||||||
|
let body = receiver.recv().unwrap();
|
||||||
|
let body = collect_body(body).await;
|
||||||
|
let expected_body = write_ipc_stream(&batch);
|
||||||
|
assert_eq!(&body, &expected_body);
|
||||||
|
}
|
||||||
|
|
||||||
|
#[tokio::test]
|
||||||
|
async fn test_delete() {
|
||||||
|
let table = Table::new_with_handler("my_table", |request| {
|
||||||
|
assert_eq!(request.method(), "POST");
|
||||||
|
assert_eq!(request.url().path(), "/table/my_table/delete/");
|
||||||
|
|
||||||
|
let body = request.body().unwrap().as_bytes().unwrap();
|
||||||
|
let body: serde_json::Value = serde_json::from_slice(body).unwrap();
|
||||||
|
let predicate = body.get("predicate").unwrap().as_str().unwrap();
|
||||||
|
assert_eq!(predicate, "id in (1, 2, 3)");
|
||||||
|
|
||||||
|
http::Response::builder().status(200).body("").unwrap()
|
||||||
|
});
|
||||||
|
|
||||||
|
table.delete("id in (1, 2, 3)").await.unwrap();
|
||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|||||||
@@ -349,8 +349,9 @@ impl UpdateBuilder {
|
|||||||
self
|
self
|
||||||
}
|
}
|
||||||
|
|
||||||
/// Executes the update operation
|
/// Executes the update operation.
|
||||||
pub async fn execute(self) -> Result<()> {
|
/// Returns the number of rows that were updated.
|
||||||
|
pub async fn execute(self) -> Result<u64> {
|
||||||
if self.columns.is_empty() {
|
if self.columns.is_empty() {
|
||||||
Err(Error::InvalidInput {
|
Err(Error::InvalidInput {
|
||||||
message: "at least one column must be specified in an update operation".to_string(),
|
message: "at least one column must be specified in an update operation".to_string(),
|
||||||
@@ -396,7 +397,7 @@ pub(crate) trait TableInternal: std::fmt::Display + std::fmt::Debug + Send + Syn
|
|||||||
data: Box<dyn arrow_array::RecordBatchReader + Send>,
|
data: Box<dyn arrow_array::RecordBatchReader + Send>,
|
||||||
) -> Result<()>;
|
) -> Result<()>;
|
||||||
async fn delete(&self, predicate: &str) -> Result<()>;
|
async fn delete(&self, predicate: &str) -> Result<()>;
|
||||||
async fn update(&self, update: UpdateBuilder) -> Result<()>;
|
async fn update(&self, update: UpdateBuilder) -> Result<u64>;
|
||||||
async fn create_index(&self, index: IndexBuilder) -> Result<()>;
|
async fn create_index(&self, index: IndexBuilder) -> Result<()>;
|
||||||
async fn list_indices(&self) -> Result<Vec<IndexConfig>>;
|
async fn list_indices(&self) -> Result<Vec<IndexConfig>>;
|
||||||
async fn merge_insert(
|
async fn merge_insert(
|
||||||
@@ -428,6 +429,31 @@ pub struct Table {
|
|||||||
embedding_registry: Arc<dyn EmbeddingRegistry>,
|
embedding_registry: Arc<dyn EmbeddingRegistry>,
|
||||||
}
|
}
|
||||||
|
|
||||||
|
#[cfg(all(test, feature = "remote"))]
|
||||||
|
mod test_utils {
|
||||||
|
use super::*;
|
||||||
|
|
||||||
|
impl Table {
|
||||||
|
pub fn new_with_handler<T>(
|
||||||
|
name: impl Into<String>,
|
||||||
|
handler: impl Fn(reqwest::Request) -> http::Response<T> + Clone + Send + Sync + 'static,
|
||||||
|
) -> Self
|
||||||
|
where
|
||||||
|
T: Into<reqwest::Body>,
|
||||||
|
{
|
||||||
|
let inner = Arc::new(crate::remote::table::RemoteTable::new_mock(
|
||||||
|
name.into(),
|
||||||
|
handler,
|
||||||
|
));
|
||||||
|
Self {
|
||||||
|
inner,
|
||||||
|
// Registry is unused.
|
||||||
|
embedding_registry: Arc::new(MemoryRegistry::new()),
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
}
|
||||||
|
|
||||||
impl std::fmt::Display for Table {
|
impl std::fmt::Display for Table {
|
||||||
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
fn fmt(&self, f: &mut std::fmt::Formatter<'_>) -> std::fmt::Result {
|
||||||
write!(f, "{}", self.inner)
|
write!(f, "{}", self.inner)
|
||||||
@@ -1757,9 +1783,6 @@ impl TableInternal for NativeTable {
|
|||||||
let data =
|
let data =
|
||||||
MaybeEmbedded::try_new(data, self.table_definition().await?, add.embedding_registry)?;
|
MaybeEmbedded::try_new(data, self.table_definition().await?, add.embedding_registry)?;
|
||||||
|
|
||||||
// Still use the legacy lance format (v1) by default.
|
|
||||||
// We don't want to accidentally switch to v2 format during an add operation.
|
|
||||||
// If the table is already v2 this won't have any effect.
|
|
||||||
let mut lance_params = add.write_options.lance_write_params.unwrap_or(WriteParams {
|
let mut lance_params = add.write_options.lance_write_params.unwrap_or(WriteParams {
|
||||||
mode: match add.mode {
|
mode: match add.mode {
|
||||||
AddDataMode::Append => WriteMode::Append,
|
AddDataMode::Append => WriteMode::Append,
|
||||||
@@ -1821,7 +1844,7 @@ impl TableInternal for NativeTable {
|
|||||||
}
|
}
|
||||||
}
|
}
|
||||||
|
|
||||||
async fn update(&self, update: UpdateBuilder) -> Result<()> {
|
async fn update(&self, update: UpdateBuilder) -> Result<u64> {
|
||||||
let dataset = self.dataset.get().await?.clone();
|
let dataset = self.dataset.get().await?.clone();
|
||||||
let mut builder = LanceUpdateBuilder::new(Arc::new(dataset));
|
let mut builder = LanceUpdateBuilder::new(Arc::new(dataset));
|
||||||
if let Some(predicate) = update.filter {
|
if let Some(predicate) = update.filter {
|
||||||
@@ -1833,9 +1856,11 @@ impl TableInternal for NativeTable {
|
|||||||
}
|
}
|
||||||
|
|
||||||
let operation = builder.build()?;
|
let operation = builder.build()?;
|
||||||
let ds = operation.execute().await?;
|
let res = operation.execute().await?;
|
||||||
self.dataset.set_latest(ds.as_ref().clone()).await;
|
self.dataset
|
||||||
Ok(())
|
.set_latest(res.new_dataset.as_ref().clone())
|
||||||
|
.await;
|
||||||
|
Ok(res.rows_updated)
|
||||||
}
|
}
|
||||||
|
|
||||||
async fn build_plan(
|
async fn build_plan(
|
||||||
|
|||||||
@@ -26,12 +26,12 @@ use super::TableInternal;
|
|||||||
#[derive(Debug, Clone)]
|
#[derive(Debug, Clone)]
|
||||||
pub struct MergeInsertBuilder {
|
pub struct MergeInsertBuilder {
|
||||||
table: Arc<dyn TableInternal>,
|
table: Arc<dyn TableInternal>,
|
||||||
pub(super) on: Vec<String>,
|
pub(crate) on: Vec<String>,
|
||||||
pub(super) when_matched_update_all: bool,
|
pub(crate) when_matched_update_all: bool,
|
||||||
pub(super) when_matched_update_all_filt: Option<String>,
|
pub(crate) when_matched_update_all_filt: Option<String>,
|
||||||
pub(super) when_not_matched_insert_all: bool,
|
pub(crate) when_not_matched_insert_all: bool,
|
||||||
pub(super) when_not_matched_by_source_delete: bool,
|
pub(crate) when_not_matched_by_source_delete: bool,
|
||||||
pub(super) when_not_matched_by_source_delete_filt: Option<String>,
|
pub(crate) when_not_matched_by_source_delete_filt: Option<String>,
|
||||||
}
|
}
|
||||||
|
|
||||||
impl MergeInsertBuilder {
|
impl MergeInsertBuilder {
|
||||||
|
|||||||
Reference in New Issue
Block a user