This PR makes the following aesthetic and content updates to the docs. - [x] Fix max width issue on mobile: Content should now render more cleanly and be more readable on smaller devices - [x] Improve image quality of flowchart in data management page - [x] Fix syntax highlighting in text at the bottom of the IVF-PQ concepts page - [x] Add example of Polars LazyFrames to docs (Integrations) - [x] Add example of adding data to tables using Polars (guides)
7.2 KiB
Understanding LanceDB's IVF-PQ index
An ANN (Approximate Nearest Neighbors) index is a data structure that represents data in a way that makes it more efficient to search and retrieve. Using an ANN index is faster, but less accurate than kNN or brute force search because, in essence, the index is a lossy representation of the data.
LanceDB is fundamentally different from other vector databases in that it is built on top of Lance, an open-source columnar data format designed for performant ML workloads and fast random access. Due to the design of Lance, LanceDB's indexing philosophy adopts a primarily disk-based indexing philosophy.
IVF-PQ
IVF-PQ is a composite index that combines inverted file index (IVF) and product quantization (PQ). The implementation in LanceDB provides several parameters to fine-tune the index's size, query throughput, latency and recall, which are described later in this section.
Product quantization
Quantization is a compression technique used to reduce the dimensionality of an embedding to speed up search.
Product quantization (PQ) works by dividing a large, high-dimensional vector of size into equally sized subvectors. Each subvector is assigned a "reproduction value" that maps to the nearest centroid of points for that subvector. The reproduction values are then assigned to a codebook using unique IDs, which can be used to reconstruct the original vector.
It's important to remember that quantization is a lossy process, i.e., the reconstructed vector is not identical to the original vector. This results in a trade-off between the size of the index and the accuracy of the search results.
As an example, consider starting with 128-dimensional vector consisting of 32-bit floats. Quantizing it to an 8-bit integer vector with 4 dimensions as in the image above, we can significantly reduce memory requirements.
!!! example "Effect of quantization"
Original: `128 × 32 = 4096` bits
Quantized: `4 × 8 = 32` bits
Quantization results in a **128x** reduction in memory requirements for each vector in the index, which is substantial.
Inverted file index
While PQ helps with reducing the size of the index, IVF primarily addresses search performance. The primary purpose of an inverted file index is to facilitate rapid and effective nearest neighbor search by narrowing down the search space.
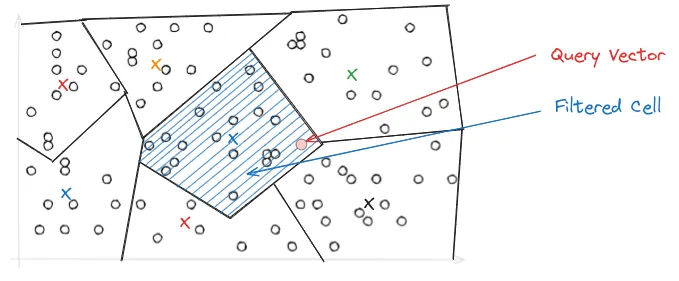
In IVF, the PQ vector space is divided into Voronoi cells, which are essentially partitions that consist of all the points in the space that are within a threshold distance of the given region's seed point. These seed points are used to create an inverted index that correlates each centroid with a list of vectors in the space, allowing a search to be restricted to just a subset of vectors in the index.
During query time, depending on where the query lands in vector space, it may be close to the border of multiple Voronoi cells, which could make the top-k results ambiguous and span across multiple cells. To address this, the IVF-PQ introduces the nprobe parameter, which controls the number of Voronoi cells to search during a query. The higher the nprobe, the more accurate the results, but the slower the query.
Putting it all together
We can combine the above concepts to understand how to build and query an IVF-PQ index in LanceDB.
Construct index
There are three key parameters to set when constructing an IVF-PQ index:
metric: Use anL2euclidean distance metric. We also supportdotandcosinedistance.num_partitions: The number of partitions in the IVF portion of the index.num_sub_vectors: The number of sub-vectors that will be created during Product Quantization (PQ).
In Python, the index can be created as follows:
# Create and train the index for a 1536-dimensional vector
# Make sure you have enough data in the table for an effective training step
tbl.create_index(metric="L2", num_partitions=256, num_sub_vectors=96)
The num_partitions is usually chosen to target a particular number of vectors per partition. num_sub_vectors is typically chosen based on the desired recall and the dimensionality of the vector. See the FAQs below for best practices on choosing these parameters.
Query the index
# Search using a random 1536-dimensional embedding
tbl.search(np.random.random((1536))) \
.limit(2) \
.nprobes(20) \
.refine_factor(10) \
.to_pandas()
The above query will perform a search on the table tbl using the given query vector, with the following parameters:
limit: The number of results to returnnprobes: The number of probes determines the distribution of vector space. While a higher number enhances search accuracy, it also results in slower performance. Typically, settingnprobesto cover 5–10% of the dataset proves effective in achieving high recall with minimal latency.refine_factor: Refine the results by reading extra elements and re-ranking them in memory. A higher number makes the search more accurate but also slower (see the FAQ page for more details on this).to_pandas(): Convert the results to a pandas DataFrame
And there you have it! You now understand what an IVF-PQ index is, and how to create and query it in LanceDB.
FAQ
When is it necessary to create a vector index?
LanceDB has manually-tuned SIMD code for computing vector distances. In our benchmarks, computing 100K pairs of 1K dimension vectors takes <20ms. For small datasets (<100K rows) or applications that can accept up to 100ms latency, vector indices are usually not necessary.
For large-scale or higher dimension vectors, it is beneficial to create vector index.
How big is my index, and how much memory will it take?
In LanceDB, all vector indices are disk-based, meaning that when responding to a vector query, only the relevant pages from the index file are loaded from disk and cached in memory. Additionally, each sub-vector is usually encoded into 1 byte PQ code.
For example, with 1024-dimension vectors, if we choose num_sub_vectors = 64, each sub-vector has 1024 / 64 = 16 float32 numbers. Product quantization can lead to approximately 16 * sizeof(float32) / 1 = 64 times of space reduction.
How to choose num_partitions and num_sub_vectors for IVF_PQ index?
num_partitions is used to decide how many partitions the first level IVF index uses. Higher number of partitions could lead to more efficient I/O during queries and better accuracy, but it takes much more time to train. On SIFT-1M dataset, our benchmark shows that keeping each partition 1K-4K rows lead to a good latency/recall.
num_sub_vectors specifies how many PQ short codes to generate on each vector. Because PQ is a lossy compression of the original vector, a higher num_sub_vectors usually results in less space distortion, and thus yields better accuracy. However, a higher num_sub_vectors also causes heavier I/O and more PQ computation, and thus, higher latency. dimension / num_sub_vectors should be a multiple of 8 for optimum SIMD efficiency.